Tensorflow implementation of the V-Net architecture for medical imaging segmentation.
This is a Tensorflow implementation of the V-Net architecture used for 3D medical imaging segmentation. This code adopts the tensorflow graph from https://github.com/MiguelMonteiro/VNet-Tensorflow. The repository covers training, evaluation and prediction modules for the (multimodal) 3D medical image segmentation in multiple classes.
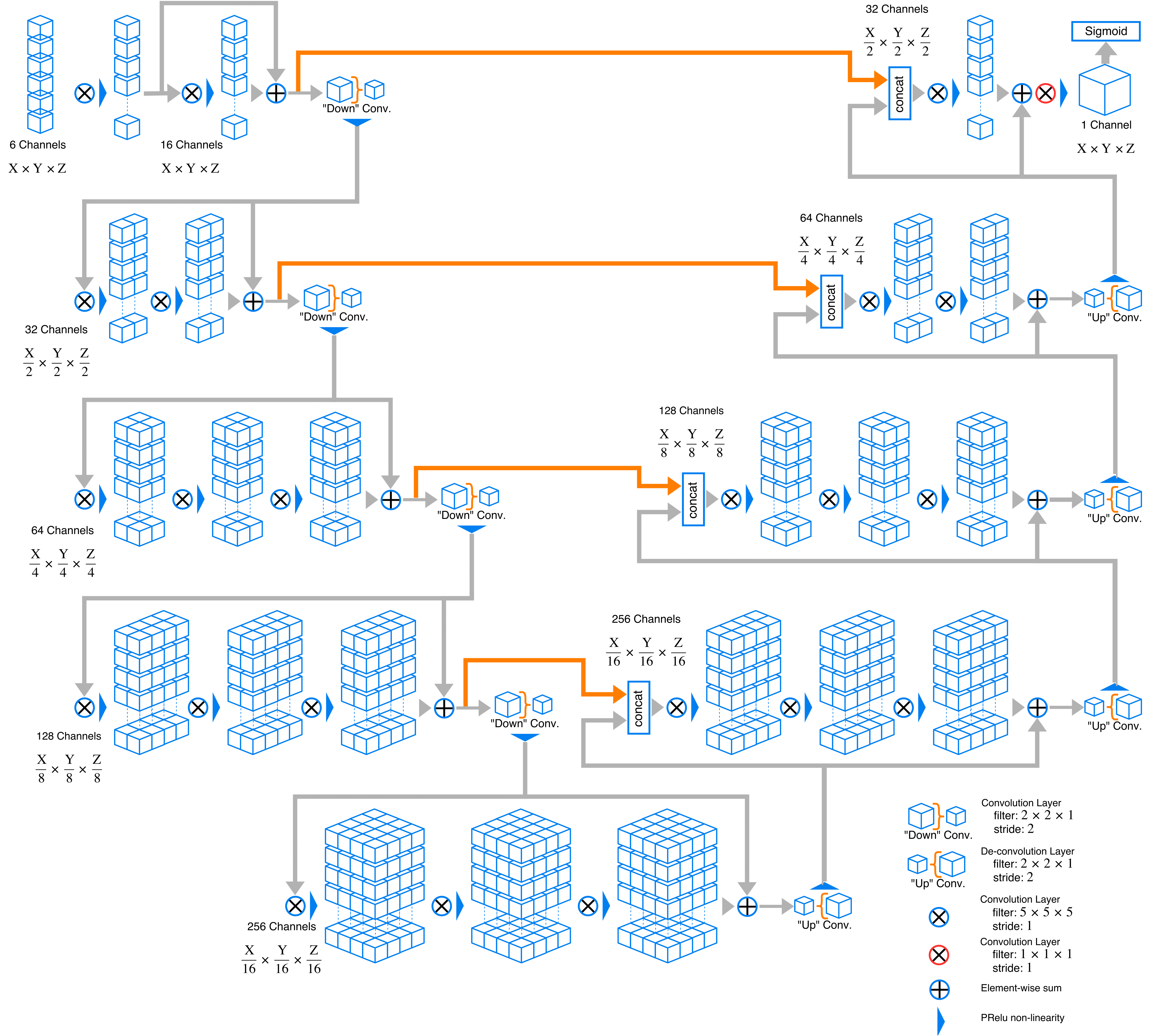
Here is an example graph of network this code implements. Channel depth may change owning to change in modality number and class number.
- 3D data processing ready
- Augumented patching technique, requires less image input for training
- Multichannel input and multiclass output
- Generic image reader with Nibabel support (Currently only support .nii/.nii.gz format for convenience, easy to expand to DICOM, tiff and jpg format)
- Medical image pre-post processing with Nibabel
- Easy network replacement structure
- Sørensen and Jaccard similarity measurement as golden standard in medical image segmentation benchmarking
- Utilizing medical image headers to retrive space and orientation info after passthrough the network
- Training
- Tensorboard visualization and logging
- Resume training from checkpoint
- Epoch training
- Evaluation from single data
- Multichannel input
- Multiclass output
- Python 3.5
- TensorFlow 1.8
- Nibabel
- mvloader
All training, testing and evaluation data should put in ./data
.
├── ...
├── data # All data
│ ├── testing # Put all testing data here
| | ├── case1
| | | ├── img.nii.gz # Image for testing
| | | └── label.nii.gz # Corresponding label for testing
| | ├── case2
| | ├──...
│ ├── training # Put all training data here
| | ├── case1 # foldername for the cases is arbitary
| | | ├── img.nii.gz # Image for testing
| | | └── label.nii.gz # Corresponding label for training
| | ├── case2
| | ├──...
│ └── evaluation # Put all evaluation data here
| | ├── case1 # foldername for the cases is arbitary
| | | └── img.nii.gz # Image for evaluation
| | ├── case2
| | ├──...
├── tmp
| ├── ckpt # Tensorflow checkpoints
| └── log # Tensorboard logging folder
├── ...
image_filename = 'img.nii(.gz)'
label_filename = 'label.nii(.gz)'In segmentation tasks, image and label are always in pair, missing either one would terminate the training process.
You may run Training.py with commandline arguments. To check usage, type python Training.py -h in terminal to list all possible training parameters.
Available training parameters
--data_location: Relative location of data folder insider working directory.
(default: './files')
(string)
--train_folder: Directory of training data.
(default: './train')
(string)
--test_folder: Directory of test data.
(default: './test')
(string)
--files: Name of files to include.
(default: ['t2_pp.nii','pd_pp.nii','mprage_pp.nii','flair_pp.nii'])
(list of strings)
--masks: Name of masks to include.
(default: ['mask.nii'])
(list of strings)
--nclasses: Number of classes.
(default: 2)
(integer)
--w: Subvolume size.
(default: [80, 80, 80])
(list of integers)
--p: Padding size.
(default: [5, 5, 5])
(list of integers)
--iterations: Number of iterations for training
(default: '10000')
(integer)
--params: List of data augmentation methods with settings (['method', setting]), e.g. ['mirror', [0,1,2]].
(default: None)
(list of strings)
--drop_out: DropOut probability.
(default: 0.5)
(float)
--loss_function: Objective function to optimize ('xent', 'sorensen', 'jaccard')
(default: 'jaccard')
(string)
--optimizer: Optimizer ('sgd', 'adam', 'momentum', 'nesterov_momentum').
(default: 'adam')
(string)
--test_each: Validation frequency in iterations.
(default: 2000)
(integer)
--checkpoint_dir: Directory where to write checkpoint
(default: './tmp/ckpt')
--decay_factor: Exponential decay learning rate factor
(default: '0.01')
(a number) (not implemented)
--decay_steps: Number of epoch before applying one learning rate decay
(default: '100')
(an integer) (not implemented)
--display_step: Display and logging interval (train steps)
(default: '1000')
(integer)
--init_learning_rate: Initial learning rate
(default: '0.1')
(float)
--save_interval: Checkpoint save interval (iterations)
(default: '1000')
(integer)
--shuffle_buffer_size: Number of elements used in shuffle buffer
(default: '5')
(integer)What the class does is basically random sampling within a image volume with provided size and padding. To this volume subsample, Gaussian-filtering can be applied and the original images together with the Gaussian-filtered versions are stacked in the channel dimension. Next, transformation are applied, if requested. The output is a tensor of shape (batch_size, nx, ny, nz, n_channels). Same holds for the masks. In addition, it is possible to selectively sample, i.e. that each n-th sample includes labelled data (by which the selectively sampled class can be determined). Read more about the inputs below.
Be aware that each class in the masks will correspond to one channel, i.e. for two classes there will be two channels (and not one).
Optional params inputs can be provided as list.
whiten: perform Gaussian-filtering of images as type bool (default: True)
subtractGaussSigma: standard deviation for Gaussian filtering as list of len 1 or ndims (default: [5])
nooriginal: use only Gaussian-filtered images as type bool (default: False)
each_with_labels: input of type int to fix the selective sampling interval, i.e. each n-th sample (default: 0, i.e. off)
minlabel: input of type int to fix which label/class to selectively sample (default: 1) deform: deformation grid spacing in voxels as list of len 1 or ndims with types int (default: [0])
deformSigma: given a deformation grid spacing, this determines the standard deviations for each dimension of the random deformation vectors as list with length 1 or ndims with types float (default: [0])
mirror: list input of len 1 or ndims of type bool to activate random mirroring along the specified axes during training (default: [0])
rotation: list input of len 1 or ndims of type float as amount in radians to randomly rotate the input around a randomly drawn vector (default: [0])
scaling: list input of len 1 or ndims of type float as amount ot randomly scale images, per dimension, or for all dimensions, as a factor, e.g. 1.25 (default: [0])
shift: list input of len 1 or ndims of type int in order to sample outside of discrete coordinates, this can be set to 1 on the relevant axes (default: [0])In training stage, result can be visualized via Tensorboard. Run the following command:
tensorboard --logdir=./tmp/logOnce TensorBoard is running, navigate your web browser to localhost:6006 to view the TensorBoard.
Note: localhost may need to change to localhost name by your own in newer version of Tensorboard.