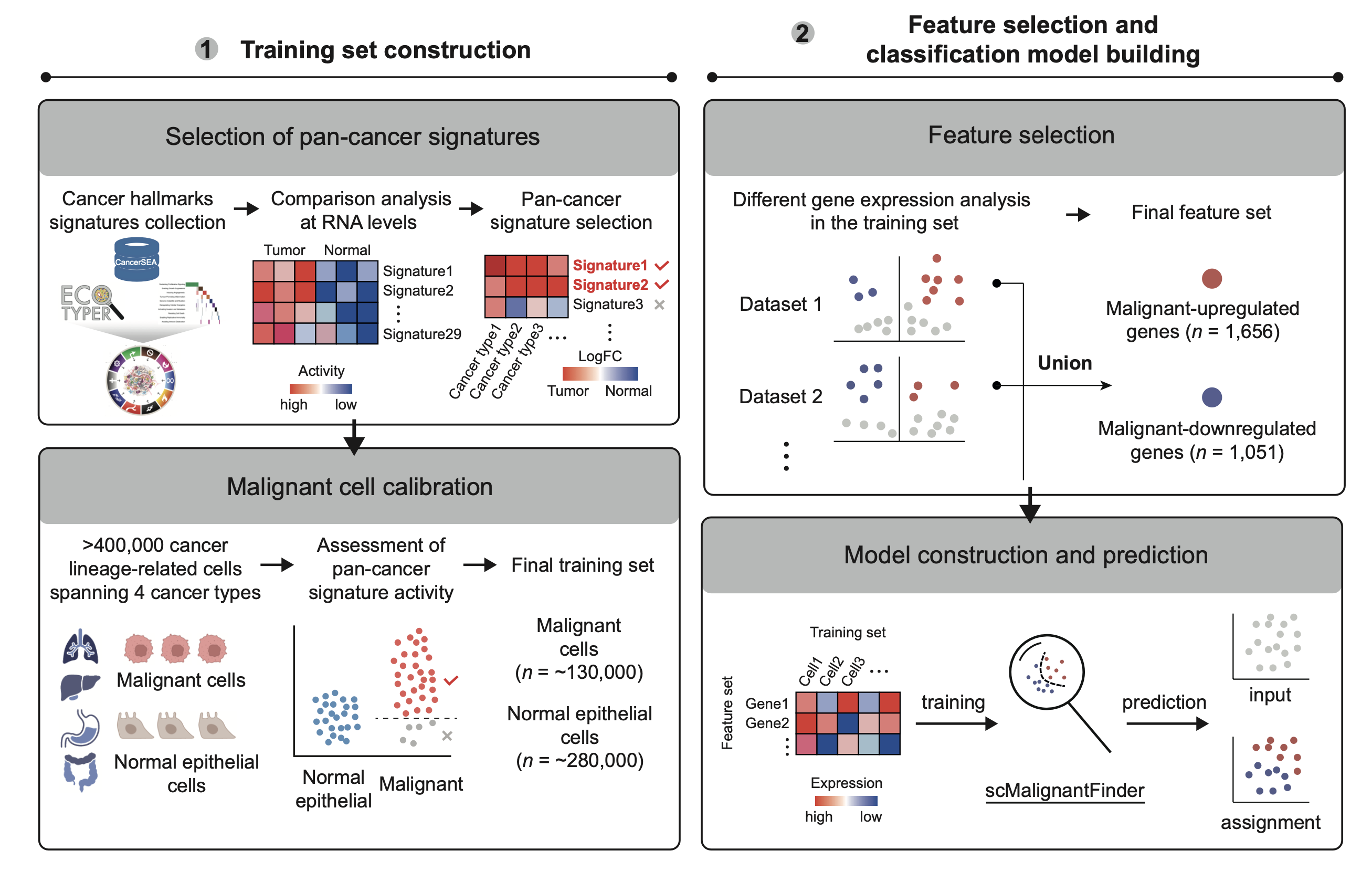
scMalignantFinder is a Python package designed for analyzing cancer single-cell RNA-seq datasets to distinguish malignant cells from their normal counterparts. Trained on over 400,000 high-quality single-cell transcriptomes, scMalignantFinder uses curated pan-cancer gene signatures for calibration and selects features by taking the union of differentially expressed genes across each dataset. For more details, please refer to the corresponding publication.
We recommend using a conda environment to install scMalignantFinder.
- Create and activate a conda environment
conda create -n scmalignant python=3.10.10
conda activate scmalignant- Install
scMalignantFinderfrom PyPI:
pip install scMalignantFinderOptional: scMalignantFinder includes a built-in pan-cancer cell type annotation tool, scATOMIC. If you want to perform basic cell type annotation before identifying malignant cells, follow the scATOMIC official tutorial to complete its installation in the same conda environment.
A pretrained model and a list of ordered features are provided in the model directory. Users can also download or use the training data for training the model.
- Training data: Download the training data used in the original study from here, or use your own dataset to train the model.
- Feature file: The feature list file can be collected from here.
- Example test data:
### Load package
from scMalignantFinder import classifier
# Initialize model
model = classifier.scMalignantFinder(
pretrain_path=None # Set the pretrain directory if you want to use the pretrained model.
train_h5ad_path='/path/to/training_data.h5ad',
feature_path='/path/to/feature_list',
test_h5ad_path='/path/to/test_data.h5ad',
celltype_annotation=False)
# celltype_annotation: If False, the cell type annotation process will not be performed. If True, use scAtomic for cell type annotation.
# Model prediction
features = model.fit()
test_adata = model.predict(features)
# View prediction
print(test_adata.obs['scMalignantFinder_prediction'].head())
# Output example:
## Index
## KUL01-T_AAACCTGGTCTTTCAT Tumor
## KUL01-T_AAACGGGTCGGTTAAC Tumor
## KUL01-T_AAAGATGGTATAGGGC Normal
## KUL01-T_AAAGATGGTGGCCCTA Tumor
## KUL01-T_AAAGCAAGTAAACACA Tumor
## Name: scMalignantFinder_prediction, dtype: category
## Categories (2, object): ['Tumor', 'Normal']If you use scMalignantFinder in your research, please cite the corresponding publication.