This package allows for importing most common motif types into R for use by functions provided by other Bioconductor motif-related packages. Motifs can be exported into most major motif formats from various classes as defined by other Bioconductor packages. Furthermore, this package allows for easy manipulation of motifs, such as creation, trimming, shuffling, P-value calculations, filtering, type conversion, reverse complementation, alphabet switching, random motif site generation, and comparison. Alongside are also included functions for interacting with sequences, such as motif scanning and enrichment, as well as sequence creation and shuffling functions. Finally, this package implements higher-order motifs, allowing for more accurate sequence scanning and motif enrichment.
if (!requireNamespace("BiocManager", quietly=TRUE))
install.packages("BiocManager")
BiocManager::install("universalmotif")if (!requireNamespace("BiocManager", quietly=TRUE))
install.packages("BiocManager")
BiocManager::install("bjmt/universalmotif")Note: building the vignettes when installing from source is not recommended, unless you don't mind waiting an hour for the necessary dependencies to install.
All of the functions within the universalmotif package are fairly well documented. You can access it from within R, reading the Bioconductor PDF, or browsing the rdrr.io website. Additionally, several vignettes come with the package, which you can access from within R or on the Bioconductor website:
- Introduction to sequence motifs
- Motif import, export, and manipulation
- Sequence manipulation and scanning
- Motif comparisons and P-values
A few key functions are also explored below.
The universalmotif class is used to store the motif matrix itself, as well as other basic information such as alphabet, background frequencies, strand, and various other metadata slots. There are a number of ways of getting universalmotif class motifs:
- Manual motif creation with
create_motif()using one of several possible input types:- Consensus sequence
- Sequence sites
- Numeric matrix
- No input: generate random motifs of any length
universalmotif class motifs are highly interoperable with other motif formats:
- Import/export from/to several supported formats:
CIS-BP:read_cisbp()HOMER:read_homer(),write_homer()JASPAR:read_jaspar(),write_jaspar()MEME:read_meme(),write_meme()TRANSFAC:read_transfac(),write_transfac()UNIPROBE:read_uniprobe()- Generic matrices:
read_matrix(),write_matrix()
- Conversion from/to several compatible Bioconductor package motif classes using
convert_motifs()(some formats cannot go both ways; see the documentation for details):TFBSTools:PFMatrix,PWMatrix,ICMatrix,PFMatrixList,PWMatrixList,ICMatrixList,TFFMFirstMotifDb:MotifListseqLogo:pwmmotifStack:pcm,pfmPWMEnrich:PWMmotifRG:MotifBiostrings:PWMrGADEM:motif
library(universalmotif)
create_motif()
#>
#> Motif name: motif
#> Alphabet: DNA
#> Type: PPM
#> Strands: +-
#> Total IC: 11.46
#> Consensus: YGTGMMMRGA
#>
#> Y G T G M M M R G A
#> A 0.17 0 0.00 0.04 0.58 0.62 0.29 0.47 0.08 0.77
#> C 0.36 0 0.01 0.00 0.41 0.36 0.68 0.16 0.05 0.00
#> G 0.00 1 0.03 0.95 0.00 0.00 0.04 0.28 0.86 0.23
#> T 0.47 0 0.96 0.02 0.00 0.03 0.00 0.09 0.00 0.00An important aspect of motif scanning and enrichment is to compare the results with those from a set of random or background sequences. For this, two functions are provided:
create_sequences(): create sequences of any alphabet, with optional desired background frequenciesshuffle_sequences(): shuffle a set of sequences, preserving any size k-let
library(universalmotif)
seqs <- create_sequences()
seqs
#> A DNAStringSet instance of length 100
#> width seq
#> [1] 100 AGTACGTTCGCATGGCAGGCATTATTTGCGCTG...TATCAGCCTAGAAGCAGGCGTACCAAGGTCTA
#> [2] 100 AATATCGGGCGCGAAGCCCGATGCGTGCTCGGA...GATGCAGTTCAAACGAAATCTCGTAAACGTGA
#> [3] 100 AGTACAGCAATGGGGACATAAGCCGTCTCATCG...CATAGTTCTCGAAATATGAATCTCCAGTCCCA
#> [4] 100 CAGATGCACTATCACCGTGCCGAGCTCGGTAAC...AATCGCATTGAACTAACAGGGGAGCAAGATAA
#> [5] 100 CGGCCCCTGGGACGTTGGATCCAGATAAAGCTT...TATGTTCCTTGCCGGAATACGGCACATATCTC
#> ... ... ...
#> [96] 100 CGGTGCAAAATGTGCCGCACACGGTAGTGCGGG...TTACACGCGTCTTTCGGAGAATGAGCTCGGCA
#> [97] 100 CAGTTAATCTATTAATGAGTCACTTAGGATTCC...GTTGCTTGGATATGGGAGAGAATGGCCAGTAA
#> [98] 100 GGGTCGTTGGCAGGGATGCACACAGACACGAAT...GTTTGCAAGACAACAGTAGCTAATTGTGCCAA
#> [99] 100 GCCTTCGGACGCCAAGTCTGCAAACAATTCCTC...CTTCTACGCCAAAACTCTTATCCCTGGCATTC
#> [100] 100 GTCACAGCCAAGCTTTAAGTCTTCCAACCAGGA...ATTGTGGACGGAAGGTACCGTCGTAGATTCGC
seqs.shuffled <- shuffle_sequences(seqs, k = 3)Additionally, if you are interested in the detailed kmer content of you sequences you can use get_bkg(). It can be used to calculate sequence background for any size kmer, and for any sequence alphabet. Results can be shown for individual sequences or merged together. There is also an option to calculate these results in any size windows (with any size overlap between windows) across the sequences.
library(universalmotif)
data(ArabidopsisPromoters)
get_bkg(ArabidopsisPromoters, merge.res = FALSE)
#> DataFrame with 4200 rows and 4 columns
#> sequence klet count probability
#> <character> <character> <integer> <numeric>
#> 1 AT4G28150 A 318 0.318
#> 2 AT1G19380 A 309 0.309
#> 3 AT4G19520 A 325 0.325
#> 4 AT1G03850 A 338 0.338
#> 5 AT5G01810 A 317 0.317
#> ... ... ... ... ...
#> 4196 AT5G22690 TTT 36 0.0360721
#> 4197 AT1G05670 TTT 43 0.0430862
#> 4198 AT1G06160 TTT 56 0.0561122
#> 4199 AT5G24660 TTT 43 0.0430862
#> 4200 AT3G19200 TTT 34 0.0340681
get_bkg(ArabidopsisPromoters, window = TRUE)
#> DataFrame with 840 rows and 5 columns
#> start stop klet count probability
#> <numeric> <numeric> <character> <integer> <numeric>
#> 1 1 100 A 1604 0.3208
#> 2 101 200 A 1636 0.3272
#> 3 201 300 A 1773 0.3546
#> 4 301 400 A 1791 0.3582
#> 5 401 500 A 1716 0.3432
#> ... ... ... ... ... ...
#> 836 501 600 TTT 255 0.0520408
#> 837 601 700 TTT 269 0.0548980
#> 838 701 800 TTT 233 0.0475510
#> 839 801 900 TTT 255 0.0520408
#> 840 901 1000 TTT 271 0.0553061The universalmotif package provides the scan_sequences() function to quickly scan a set of input sequences for motif hits. Additionally, the add_multifreq() function can be used to generate higher order motifs. These can also be used to scan sequences with higher accuracy.
library(universalmotif)
library(Biostrings)
data(ArabidopsisPromoters)
seqs <- DNAStringSet(rep(c("CAAAACC", "CTTTTCC"), 3))
motif <- create_motif(seqs, pseudocount = 1)
scan_sequences(motif, ArabidopsisPromoters, threshold = 0.9,
threshold.type = "logodds")
#> DataFrame with 53 rows and 12 columns
#> motif motif.i sequence start stop score match
#> <character> <integer> <character> <integer> <integer> <numeric> <character>
#> 1 motif 1 AT4G28150 621 627 9.08 CTAAACC
#> 2 motif 1 AT1G19380 139 145 9.08 CTTATCC
#> 3 motif 1 AT1G19380 204 210 9.08 CTAAACC
#> 4 motif 1 AT1G03850 203 209 9.08 CTAATCC
#> 5 motif 1 AT5G01810 821 827 9.08 CATATCC
#> ... ... ... ... ... ... ... ...
#> 49 motif 1 AT1G19510 960 966 9.08 CTTTTCC
#> 50 motif 1 AT5G22690 81 87 9.08 CAATACC
#> 51 motif 1 AT5G22690 362 368 9.08 CAAATCC
#> 52 motif 1 AT1G06160 956 962 9.08 CTAATCC
#> 53 motif 1 AT3G19200 365 371 9.08 CATTACC
#> thresh.score min.score max.score score.pct strand
#> <numeric> <numeric> <numeric> <numeric> <character>
#> 1 8.172 -19.649 9.08 100 +
#> 2 8.172 -19.649 9.08 100 +
#> 3 8.172 -19.649 9.08 100 +
#> 4 8.172 -19.649 9.08 100 +
#> 5 8.172 -19.649 9.08 100 +
#> ... ... ... ... ... ...
#> 49 8.172 -19.649 9.08 100 +
#> 50 8.172 -19.649 9.08 100 +
#> 51 8.172 -19.649 9.08 100 +
#> 52 8.172 -19.649 9.08 100 +
#> 53 8.172 -19.649 9.08 100 +
motif.k2 <- add_multifreq(motif, seqs, add.k = 2)
scan_sequences(motif.k2, ArabidopsisPromoters, use.freq = 2, threshold = 0.9,
threshold.type = "logodds")
#> DataFrame with 8 rows and 12 columns
#> motif motif.i sequence start stop score match
#> <character> <integer> <character> <integer> <integer> <numeric> <character>
#> 1 motif 1 AT4G12690 938 943 17.827 CAAAAC
#> 2 motif 1 AT2G37950 751 756 17.827 CAAAAC
#> 3 motif 1 AT1G49840 959 964 17.827 CTTTTC
#> 4 motif 1 AT1G77210 184 189 17.827 CAAAAC
#> 5 motif 1 AT1G77210 954 959 17.827 CAAAAC
#> 6 motif 1 AT3G57640 917 922 17.827 CTTTTC
#> 7 motif 1 AT4G14365 977 982 17.827 CTTTTC
#> 8 motif 1 AT1G19510 960 965 17.827 CTTTTC
#> thresh.score min.score max.score score.pct strand
#> <numeric> <numeric> <numeric> <numeric> <character>
#> 1 16.0443 -16.842 17.827 100 +
#> 2 16.0443 -16.842 17.827 100 +
#> 3 16.0443 -16.842 17.827 100 +
#> 4 16.0443 -16.842 17.827 100 +
#> 5 16.0443 -16.842 17.827 100 +
#> 6 16.0443 -16.842 17.827 100 +
#> 7 16.0443 -16.842 17.827 100 +
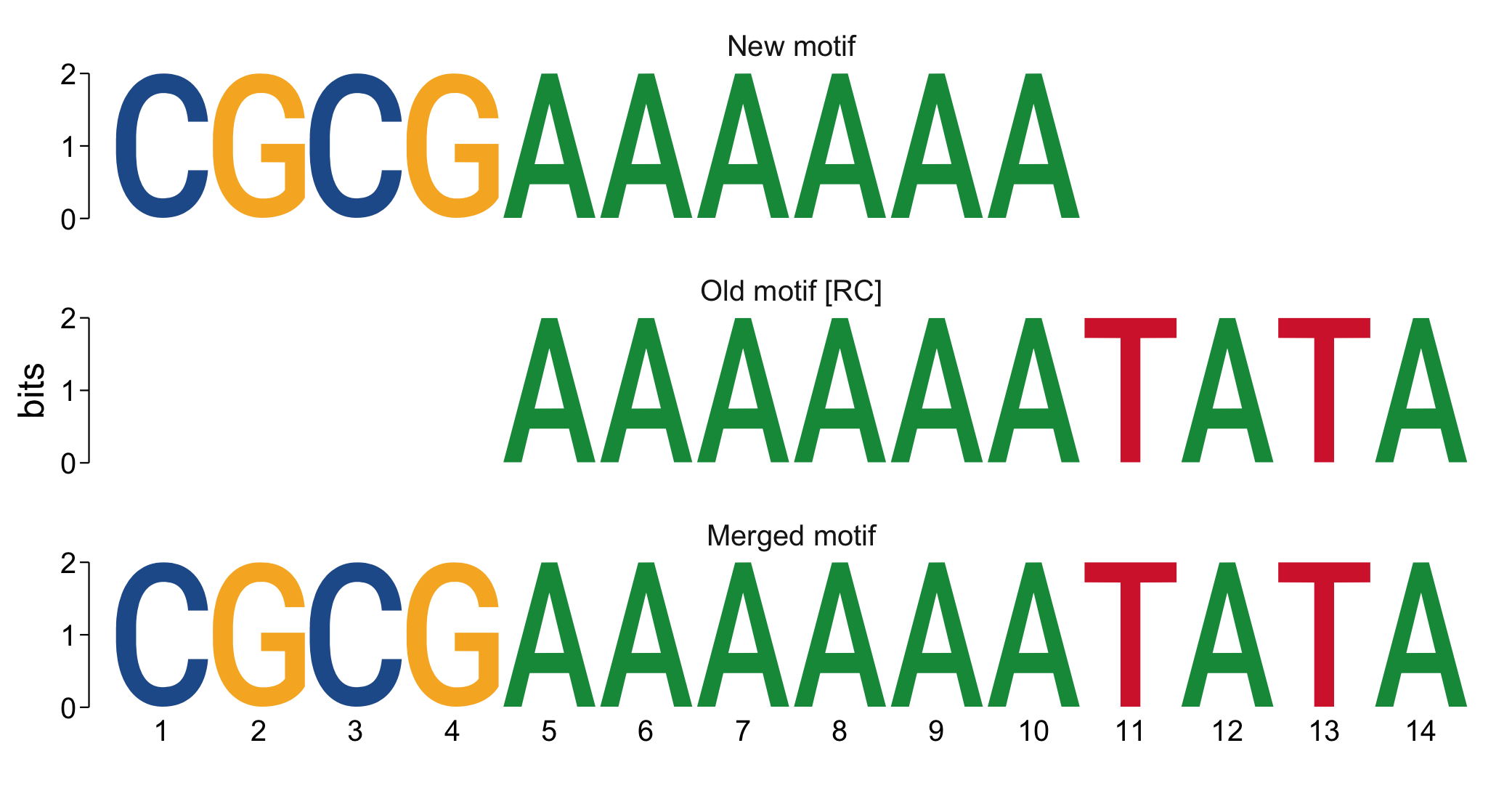
#> 8 16.0443 -16.842 17.827 100 +A commonly performed task after de novo motif discovery is to check how closely it might resemble known motifs. This can be performed using the highly customizable compare_motifs() with one of several available metrics. Different motifs can also be merged with merge_motifs().
library(universalmotif)
new.motif <- create_motif("CGCGAAAAAA", name = "New motif")
old.motif <- create_motif("TATATTTTTT", name = "Old motif")
compare_motifs(c(new.motif, old.motif), method = "PCC", min.overlap = 5)[2]
#> [1] 1
compare_motifs(c(new.motif, old.motif), method = "PCC", min.overlap = 10)[2]
#> [1] 0.2
merged.motif <- merge_motifs(c(new.motif, old.motif), method = "PCC",
new.name = "Merged motif")
view_motifs(c(new.motif, old.motif, merged.motif))