ExpansionHunter Denovo (EHdn) is a suite of tools for detecting novel expansions of short tandem repeats (STRs). EHdn is intended for analysis of a collection of BAM/CRAM files containing alignments of short (100-200bp) reads.
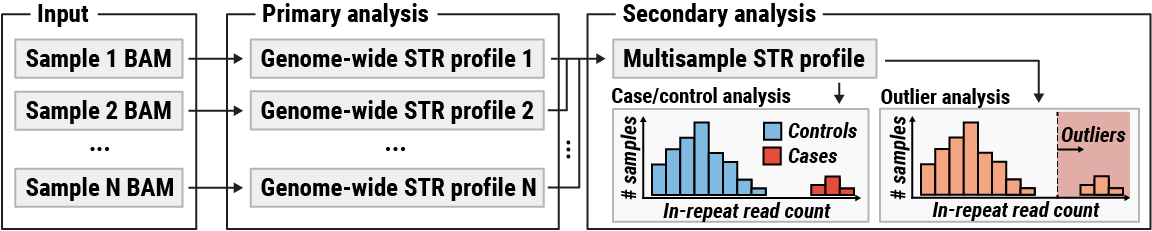
As shown in the figure above, the analysis workflow consists of two steps. During the first step, genome-wide STR profiles are extracted from the input BAM files. The STR profiles contain information about reads that originate in STRs longer than the read length. The second step involves comparing STR profiles to each other. The type of comparison depends on the dataset:
| Analysis type | Dataset |
|---|---|
| Case-control | Cases are enriched in expansions of the same STR |
| Outlier | Only a few cases are expected to contain the same STR expansion |
For example, if a case-control analysis is applied to a dataset consisting of ALS patients and healthy controls, then it is expected to flag the GGCCCC repeat in C9orf72 gene as highly significant. On the other hand, if cases consist of samples from patients with diverse phenotypes, it might be appropriate to assume that there is no enrichment for any specific expansion and hence the case-control analysis is not appropriate. In this situation, an outlier analysis can be used to flag repeats that are expanded in a small proportion of cases compared to the rest of the dataset.
- Approximate location and nucleotide composition of STRs are inferred automatically.
- A single BAM/CRAM file can be analyzed in about 30 mins to 5 hours on a typical workstation. The exact runtime will depend on the sensitivity settings.
- STRs shorter than the read length are ignored; the program is appropriate only for detecting expansions that exceed the read length.
- The location of each reported STR is approximate (up to about 500bp-1Kbp)
- STRs are not genotyped; the program reports a depth-normalized count of reads originating inside each STR; this count can be used as a very approximate measure of the repeat length
- To achieve best results all samples must be sequenced on the same instrument to similar coverage, have the same read and fragment lengths, and be subjected to the same computational pre-processing (e.g. reads must be aligned by the same aligner)
See documentation for installation instructions, usage guide, and description of file formats.
ExpansionHunter Denovo is provided under the terms and conditions of the PolyForm Strict License 1.0.0. It relies on several third party packages provided under other open-source licenses, please see COPYRIGHT.txt for additional details.