Omics Compendium Builder (OCB): An automated omics compendium preparation pipeline
This toolkit can prepare the transcriptomic compendium, a normalized, format-consistent data matrix across samples from different studies, by collecting the samples in Sequencing Read Archive (SRA) database given the topic you are interested in and your target species.
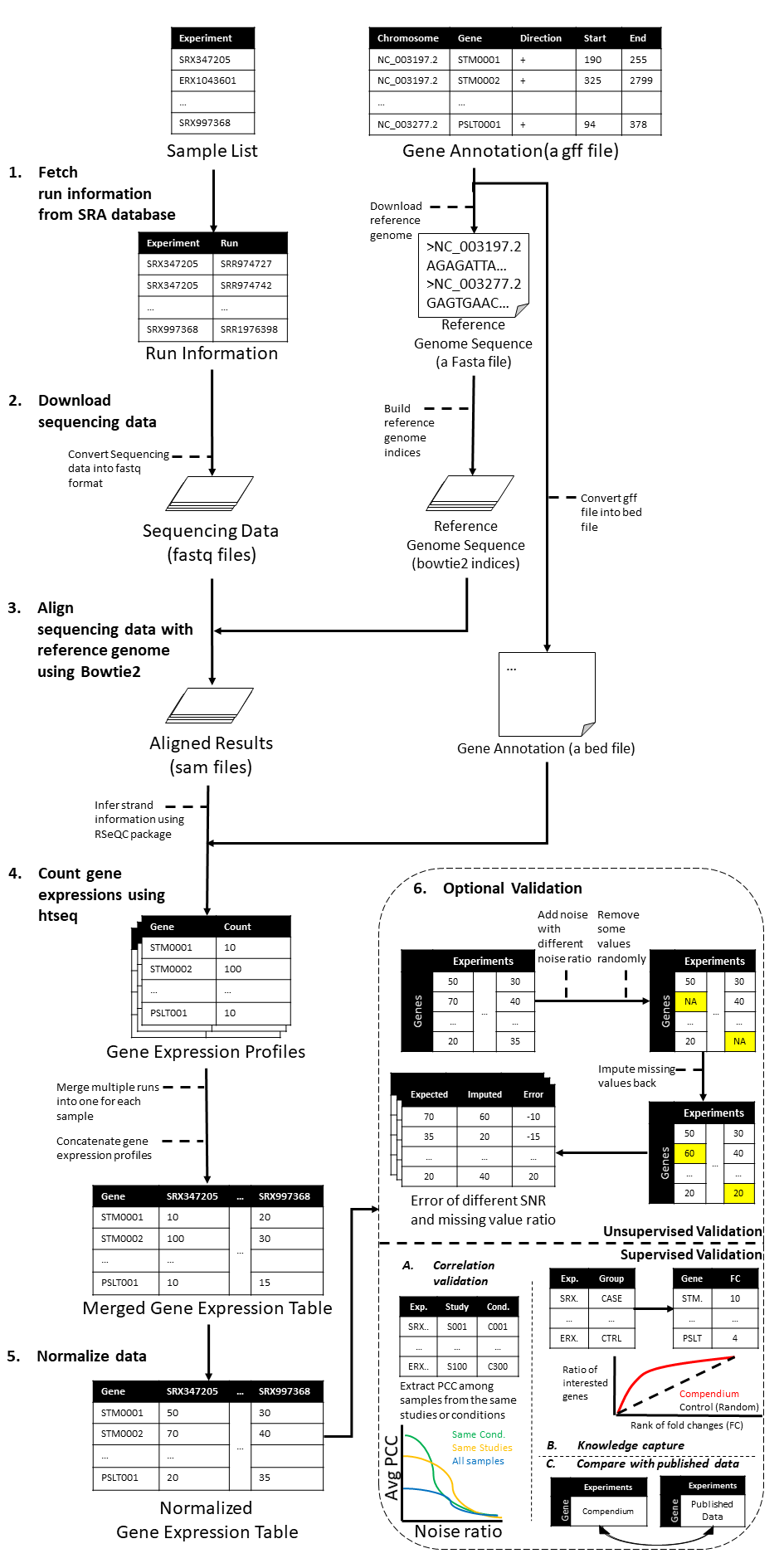 Figure 1. The entire transcriptomic compendium pipeline. The process consists of 6 steps: 1, Metadata preparation by extracting run information from SRA. 2, Downloading sequencing data in FASTA format. 3, Aligning sequences with reference genomes. 4, Generating gene expression profile for each run given the corresponding sequence direction information (BED) and gene annotation. 5, Normalizing gene expression profile table. 6, Different approaches for validating the quality of the generated compendium.
Figure 1. The entire transcriptomic compendium pipeline. The process consists of 6 steps: 1, Metadata preparation by extracting run information from SRA. 2, Downloading sequencing data in FASTA format. 3, Aligning sequences with reference genomes. 4, Generating gene expression profile for each run given the corresponding sequence direction information (BED) and gene annotation. 5, Normalizing gene expression profile table. 6, Different approaches for validating the quality of the generated compendium.
Directories
TranscriptomicPipelines: The folder contains the source code for OCB.
Getting Started
Download the entire repository:
git clone https://github.com/IBPA/OCB.git
Dependencies
For Ubuntu users, there is the script to install software and Python packages once you have installed Python3.6 and pip packages:
Adjust the file access mode if you cannot run the script, for example:
chmod 755 install.bash
./install.bash
After the script finish the installation, please follow the instuction to add the installation path to PATH variable.
Software
Make sure the following softwares are installed. The following version has been tested.
Generally, it is good to use newer version even though it is not tested, but there will be some issues if older version is used.
python==3.6.9
sra-tools==2.10.8
bowtie==2.3.4
You can use the following official links to download the software with specified version:
Then set the PATH variable so that you can call the executable everywhere.
The SRA toolkits may need additional configuration. You can run the following command to check the configuration:
prefetch --version
Run the following command, edit and save the configuration if you cannot see the version:
vdb-config -i
Packages
Make sure to install the following Python packages.
Generally it is good to use the newer packages except scikit-learn package.
biopython==1.74
pandas==0.25
RSeQC==3.0.0
HTSeq==0.11.2
missingpy==0.2.0
scikit-learn==0.20.1
matplotlib==3.0.2
Running
The pipeline consists of two components: Compendium construction and validation. The pipeline builds a compendium using the sample lists and gene annotations provided by users. Then it provides different validation approaches to validate the statistical siginificance and usefulness of the generated compendiums. For more detailed usage, see this step-by-step tutorial.
Constructing Compendium
Input
In order to build a compendium, the script needs three input arguments:
- The path to a sample list file (Example, Simple Example)
- The path to a gene annotation file.
- An output compendium name.
Output
This script will generate a directory with specified compendium name and many files in the directory. There are two outputs that are the most important:
- Normalized data matrix: A CSV table that contains normalized gene expression profiles of all samples. Each row represents different genes and each column represents different samples. The output is stored in '($compendium_name)_NormalizedDataMatrix.csv'.
- Compendium in binary format: A python object that contains the normalized gene expression table and the recorded parameters. It can be used for optional validation. The output is stored in '($compendium_name)_projectfile.bin'.
Example
This example demostrate the simple compendium construction. It can be finished less than 5 minutes using a laptop with correct configuration (NOTE: This compendium is just for demo which allow users to view the format of output files, but cannot be validated due to limited sample number and sample size)
cd TranscriptomicPipelines
python build_compendium_script.py \
../TestFiles/SimpleSalmonellaSampleList.csv \
../TestFiles/GCF_000006945.2_ASM694v2 \
SimpleSalmonellaExample
Example (Will be time consuming)
This example process most of the Salmonella RNA-seq samples available in SRA. The compendium can be validated with four different approches (see the next part), but the compendium construction will be time consuming, which takes about one week if you process eight samples in parallel in cluster. To try the validation processes, please use this [processed compendium] (./SalmonellaExample.tar.gz).
cd TranscriptomicPipelines
python build_compendium_script.py \
../TestFiles/SalmonellaExampleSampleList.csv \
../TestFiles/GCF_000006945.2_ASM694v2 \
SalmonellaExample
Validating Compendium
The pipeline provides several approaches to ensure the quality of the generated compendiums:
- Unsupervised validation
- Supervised validation with correlation
- Supervised validation with knowledge capture
- Supervised validation with published data
Please refer to validation totorial.
Authors
- ChengEn Tan as the project lead, main author, and the main developer.
- Fangzhou Li as the metadata pipeline developer and the code reviewer.
- Dr. Minseung Kim as the technical advisor.
- Dr. Ilias Tagkopoulos as the project supervisor and advisor.
Contact
For any questions, please contact us at tagkopouloslab@ucdavis.edu.
License
This project is licensed under the Apache 2.0 License - see the LICENSE file for details.