author : yanhong hong
a uncomplete work
COCOON(COlleCtions Of OrgaNoid)
(1)Query1 used to search the pmc database:
((organoid) AND cancer) NOT (organoid[Reference]) AND ("2010"[Publication Date] : "3000"[Publication Date]) Search results:9680 items.
And we download the results as xml format: pmc_result_for_organoid_full_20210225.xml .
download the summery as txt format: pmc_result_for_organoid_summary_20210225.txt .
(2)Query2 used to search the pmc database: (This query seems to be more reliable)
(organoid[Title] OR organoid[Abstract] OR organoid[Body - All Words]) AND (cancer[Title] OR cancer[Abstract] OR cancer[Body - All Words])AND ("2010"[Publication Date] : "3000"[Publication Date]) Search results:5623 items.
And we download the results as xml and txt formats: pmc_result_for_organoid_full_20210226.xml and pmc_result_for_organoid_summary_20210226.txt
generate xml: https://www.xmlviewer.org/
Tree view of the xml file.
(3)Query used to search the pubmed database:
(organoid) AND (cancer) AND ("2010"[Publication Date] : "3000"[Publication Date]) Search results:2467 items.
And we download the results as xml format: pubmed_result_for_organoid_full_20210225.xml .
download the summery as txt format: pubmed_result_for_organoid_summary_20210225.txt .
(4)Patent data
in progress....
(1) tumor type
(2) sample amount
(3) omics data type
(4) the accession number of the dataset
(5)...
(6)...
(1) search for some key words for omics data.(e.g. sra, eda, gse , geo , data availability ...). result :we get a list of matched article titles.
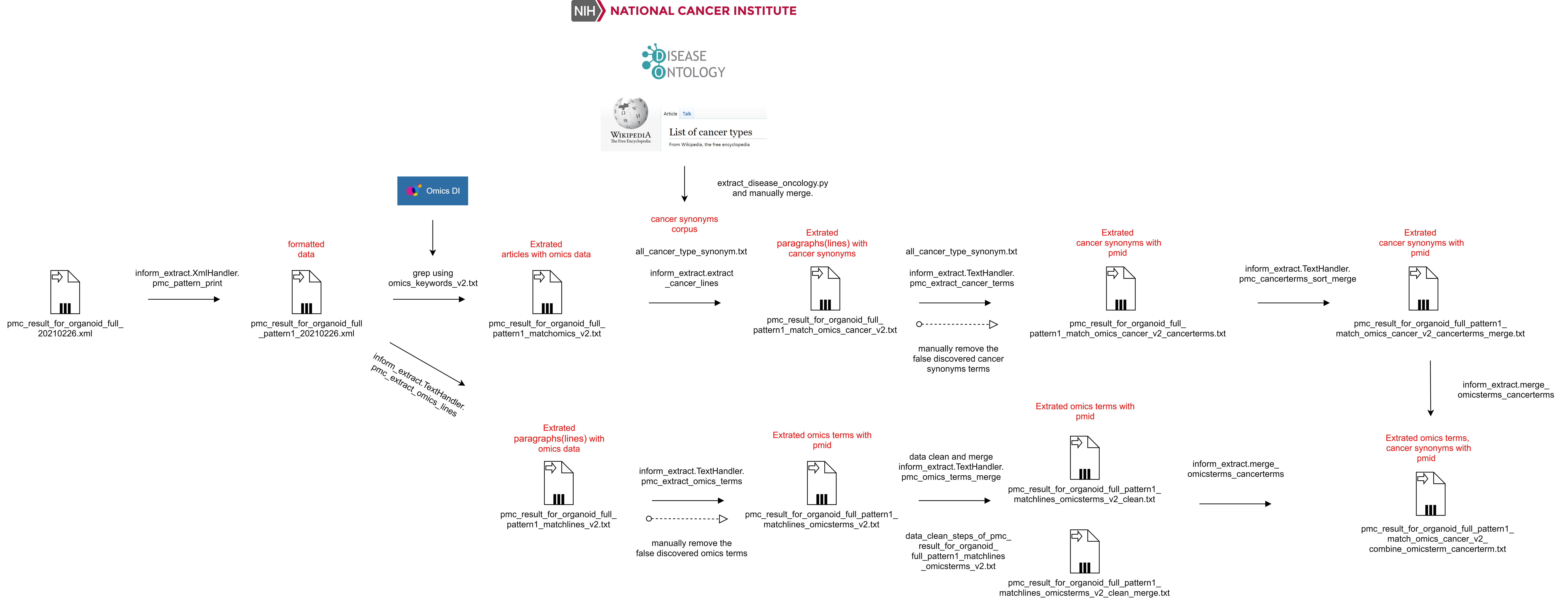
We use script inform_extract.py to rearrange the xml file to:
<article-pmid> <the article element>the result files is pmc_result_for_organoid_full_pattern1_20210226.xml
we use a list of key words about omics that we extract from the patterns of identities from different omics repository from omicsdi (https://www.omicsdi.org/database)
#omics_keywords_v2.txt
gse[[:digit:]][[:digit:]][[:digit:]]+
SRR[[:digit:]][[:digit:]][[:digit:]]+
srx[[:digit:]][[:digit:]][[:digit:]]+
srp[[:digit:]][[:digit:]][[:digit:]]+
ST[[:digit:]][[:digit:]][[:digit:]]+
PRJEB[[:digit:]][[:digit:]][[:digit:]]+
ptjna[[:digit:]][[:digit:]][[:digit:]]+
MTBLS[[:digit:]][[:digit:]][[:digit:]]+
PAe[[:digit:]][[:digit:]][[:digit:]]+
E-MTAB[[:digit:]][[:digit:]][[:digit:]]+
oep[[:digit:]][[:digit:]][[:digit:]]+
samea[[:digit:]][[:digit:]][[:digit:]]+
fr-fcm[[:digit:]][[:digit:]][[:digit:]]+
e-mtab-[[:digit:]][[:digit:]][[:digit:]]+
oex[[:digit:]][[:digit:]][[:digit:]]+
gpm[[:digit:]][[:digit:]][[:digit:]]+
pxd[[:digit:]][[:digit:]][[:digit:]]+
msv[[:digit:]][[:digit:]][[:digit:]]+
ERR[[:digit:]][[:digit:]][[:digit:]]+
erx[[:digit:]][[:digit:]][[:digit:]]+
EGAD[[:digit:]][[:digit:]][[:digit:]]+
LDG-[[:digit:]][[:digit:]][[:digit:]]+
lds-[[:digit:]][[:digit:]][[:digit:]]+
prd[[:digit:]][[:digit:]][[:digit:]]+
phs[[:digit:]][[:digit:]][[:digit:]]+
BIOMD[[:digit:]][[:digit:]][[:digit:]]+
MODEL[[:digit:]][[:digit:]][[:digit:]]+
S-BSST[[:digit:]][[:digit:]][[:digit:]]+
idr[[:digit:]][[:digit:]][[:digit:]]+
nct[[:digit:]][[:digit:]][[:digit:]]+
erp[[:digit:]][[:digit:]][[:digit:]]+
dryad
\S+\.figshare\.\S+
cat pmc_result_for_organoid_full_pattern1_20210226.xml |grep -i -E -f omics_keywords.txt > pmc_result_for_organoid_full_pattern1_match_omics.txtarticles that match the omics pattern: pmc_result_for_organoid_full_pattern1_match_omics.txt
cat pmc_result_for_organoid_full_pattern1_20210226.xml |grep -i -E -f omics_keywords_v2.txt > pmc_result_for_organoid_full_pattern1_match_omics_v2.txtpmids of articles that match the omics pattern:
we then extract lines(inform_extract.py) that matches patterns to pmc_result_for_organoid_full_pattern1_matchlines_v2.xml .(This file is for manually curate)
we then extract omics terms(inform_extract.py)that matches patterns in omics_keywords_v2.txt to pmc_result_for_organoid_full_pattern1_matchlines_v2.xml to pmc_result_for_organoid_full_pattern1_matchlines_omicsterms_v2.txt.
after manually clean and automatically merge(inform_extract.py),the result file is pmc_result_for_organoid_full_pattern1_matchlines_omicsterms_v2_clean_merge.txt.
the manually clean steps are listed in data_clean_steps_of_pmc_result_for_organoid_full_pattern1_matchlines_omicsterms_v2.txt .
(2)
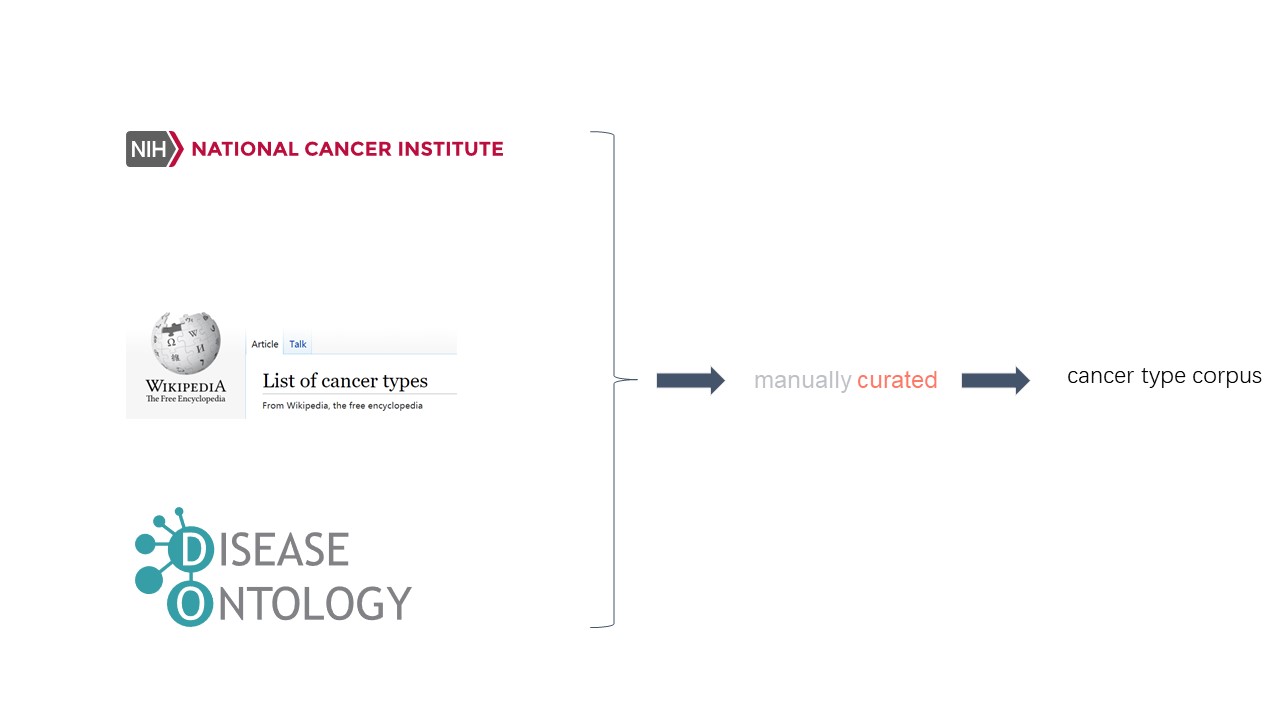
We use a list of key words (corpus) about various cancer type to extract the tumor type information from abstract and body text using the xml file format.
We extract key words from three sources: extract_disease_oncology.py .
(source 1) script1 used to extract tumor types key words from wiki.(https://en.wikipedia.org/wiki/List_of_cancer_types)
from lxml import html
from urllib.request import ProxyHandler,build_opener
proxy = ProxyHandler({'https': 'https://hyh:hyh2102011@10.10.115.225:1080'})
opener = build_opener(proxy)
url = 'https://en.wikipedia.org/wiki/List_of_cancer_types'
html_text = opener.open(url).read().decode('utf-8')
tree = html.fromstring(html_text)
elements = tree.xpath('//*[@id="mw-content-text"]/div[1]/ul/li')
for element in elements:
print ("".join(element.xpath('descendant-or-self::text()')))Manual curated the outputs. > the result file is wiki_cancer_type.txt .
(source 2) script2 used to extract tumor types key words from cancer gov.(https://www.cancer.gov/types)
import requests
from bs4 import BeautifulSoup
url = 'https://www.cancer.gov/types'
r = requests.get(url)
soup = BeautifulSoup(r.text, 'html.parser')
mydivs = soup.find_all("ul", {"class": "cancer-list"})
alltext = [row.text for row in mydivs]
for text in alltext:
print(text)Manual curated the outputs. > the result file is cancer_type_cancergov.txt .
(source 3) (the data is from https://github.com/DiseaseOntology/HumanDiseaseOntology/blob/main/src/ontology/subsets/DO_cancer_slim.json)
script used to extract key words.
result : DO_cancer_slim.json
cat cancer_type_cancergov.txt disease_oncology_lbl.txt disease_oncology_synonym.txt wiki_cancer_type.txt |sort|uniq > all_cancer_type_synonym.txtWe integrate three sources to one file: all_cancer_type_synonym.txt .
Total key words number: 1562.
e.g. remove redundancy, remove unwanted key words.
(3)
We use the pmc_result_for_organoid_full_pattern1_match_omics_v2.txt and all_cancer_type_synonym.txt as input to extract paragraphs(or lines) that has cancer synonyms,output is pmc_result_for_organoid_full_pattern1_match_omics_cancer_v2.txt .(script: inform_extract.py)
We use pmc_result_for_organoid_full_pattern1_match_omics_cancer_v2.txt and all_cancer_type_synonym.txt as input to extract cancer synonyms terms,output is pmc_result_for_organoid_full_pattern1_match_omics_cancer_v2_cancerterms.txt
the merge version is pmc_result_for_organoid_full_pattern1_match_omics_cancer_v2_cancerterms_merge.txt.
(4)
finally we combine two results pmc_result_for_organoid_full_pattern1_match_omics_cancer_v2_cancerterms_merge.txt and pmc_result_for_organoid_full_pattern1_matchlines_omicsterms_v2_clean_merge.txt to get pmc_result_for_organoid_full_pattern1_match_omics_cancer_v2_combine_omicsterm_cancerterm.txt .
(5)
the pipeline to extract information of pubmed is almost the same as pmc,the final file is pubmed_result_for_organoid_summary_pattern1_match_omics_cancer.xml.
(6)
in progress....
cases we need to consider include:
(1) The omics data comes from the organoids involved in the article.(e.g. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC6423033/ )
(2) The omics data comes from public data just for validation of the results.(e.g. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC7565382/)
try: n grams method.