This portfolio contains my notebooks about machine and deep learning algorithms applied on single-cell RNA-seq data. This data was downloaded from NeuroIPS 2021. These notebooks are described below:
1. scRNA-seq 🧬: Differential Expression with scVI (see it in action at Kaggle)
Linearly decoded VAE (LDVAE) is a generative model similar to PCA which can be used similarly as scVI, however, due that it has linear functions instead of neural networks it's more easy to interpret its ouput. In this notebook i used this model to get differential expression analysis.
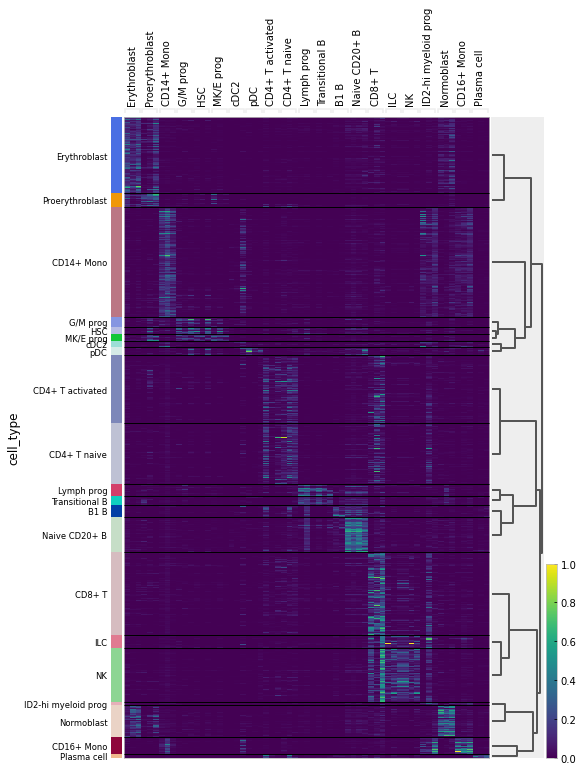
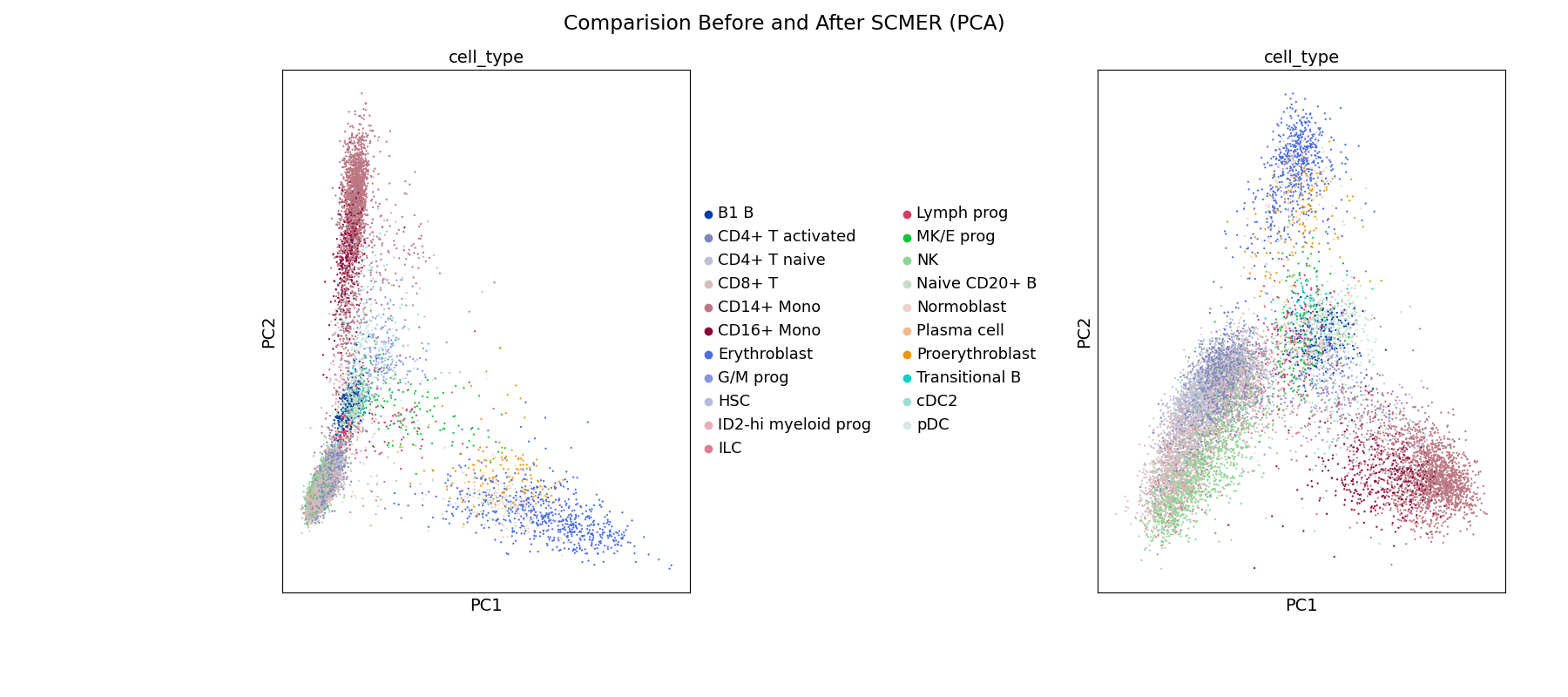
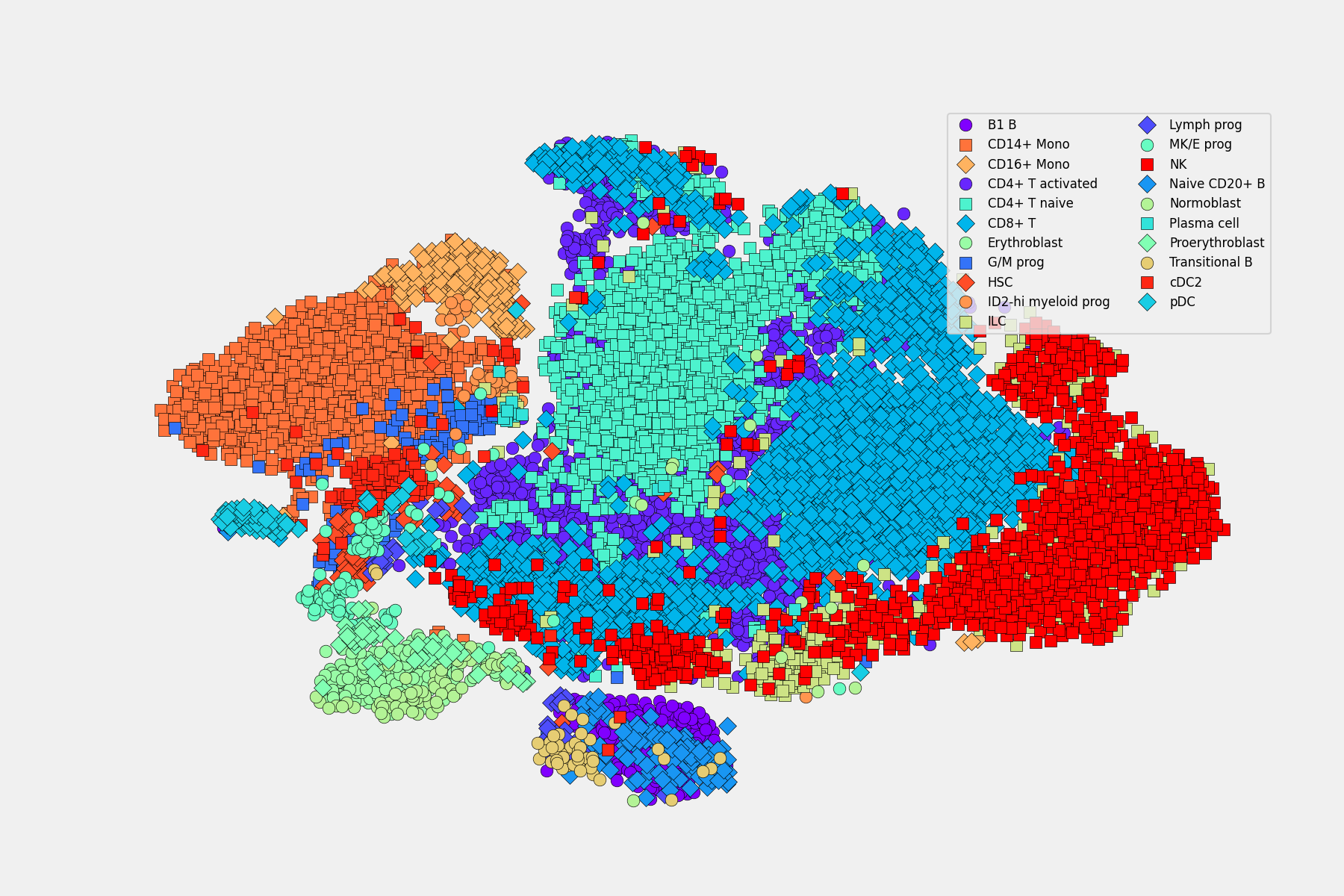
2. scRNA-seq 🧬: Scanpy & SCMER for Feature Selection (see it in action at Kaggle)
SCMER (single-cell manifold-preserving feature
selection) was used in notebook to get a compact version of the full dataset. I also do some visualizations and i have to admit that is too easy to use with Scanpy, once you have your data in form of adata object.
3. scATAC-seq 🧬: EpiScanpy & PeakVI (see it in action at Kaggle)
After some practice with SCMER i decided to use it to get a matrix that i could handle to train graph neural networks and only 13GB RAM. The GNN used is scGAE, a recently published model on Nature. scGAE performs clustering on its latent space and it can be used for downstram analysis.
Contact: hiramcoria@gmail.com