The enrichMiR R package implements various tests to identify miRNAs whose targets are enriched among genesets of interest or a differential expression signature.
To use the package internally, view the vignette. You may also use the interactive app.
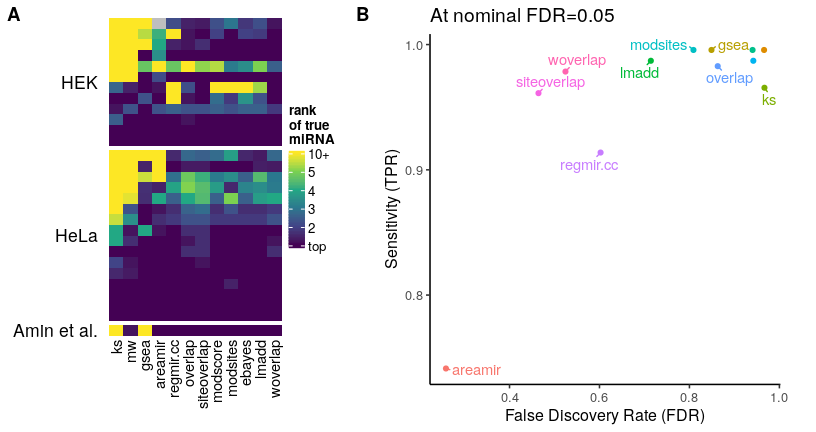
enrichMiR implements a variety of statistical tests which are described in the app and publication. To guide users, we benchmarked them using miRNA intervention experiments (partially scrambled to increase difficulty). The benchmark results are summarized in the Figure below (see publication for more detail). In short, we recommend the siteoverlap, woverlap, lmadd and areamir tests; when using large annotations (e.g. scanMiR), we recommend the lmadd test (see publication).