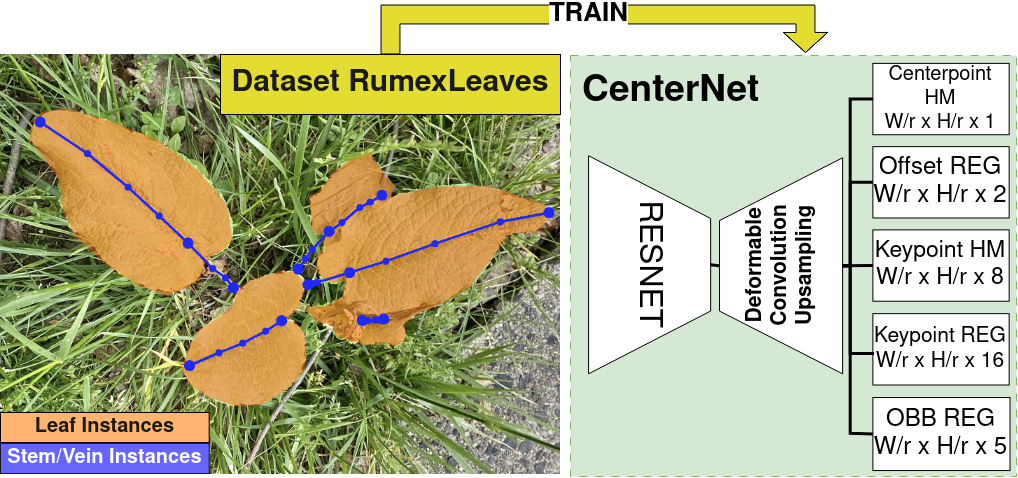
This repository contains the official implementation for
Zoom in on the Plant: Fine-grained Analysis of Leaf, Stem and Vein Instances. Ronja Güldenring, Rasmus Eckholdt Andersen and Lazaros Nalpantidis, IEEE Robotics and Automation Letters (RA-L), 2023
Abstract:
Robot perception is far from what humans are capable of. Humans do not only have a complex semantic scene understanding but also extract fine-grained intra-object properties for the salient ones. When humans look at plants, they naturally perceive the plant architecture with its individual leaves and branching system. In this work, we want to advance the granularity in plant understanding for agricultural precision robots. We develop a model to extract fine-grained phenotypic information, such as leaf-, stem-, and vein instances. The underlying dataset \textit{RumexLeaves} is made publicly available and is the first of its kind with keypoint-guided polyline annotations leading along the line from the lowest stem point along the leaf basal to the leaf apex. Furthermore, we introduce an adapted metric POKS complying with the concept of keypoint-guided polylines. In our experimental evaluation, we provide baseline results for our newly introduced dataset while showcasing the benefits of POKS over OKS.
Sources:
- Create environment and install pip requirements
make create_environment make requirements - Download & prepare data
make data - Visualize example images with annotations
conda run -n rumexleaves_centernet python rumexleaves_centernet/visualizations/visualize_data.py - Download model weights
make download_weights - Run example inference
conda run -n rumexleaves_centernet python rumexleaves_centernet/tools/inference.py --exp_file exp_files/eval_inat.py -ckpt models/final_model.pth -img data/processed/RumexLeaves/iNaturalist/4150.jpg - Run validation of final model on iNaturalist data
conda run -n rumexleaves_centernet python rumexleaves_centernet/tools/evaluate.py --exp_file exp_files/eval_inat.py -ckpt models/final_model.pth - Training final model from scratch
conda run -n rumexleaves_centernet python rumexleaves_centernet/tools/train.py --exp_file exp_files/train_final_model.py
- Download & prepare data
make data - Get submodules
git submodule update --init --recursive - Build docker image
docker build -f dockerfiles/train_model.dockerfile . -t train_model:latest - Run training in docker container
docker run --gpus all -v "$(pwd)/data/processed:/data/processed" -v "$(pwd)/log:/log" -v "$(pwd)/exp_files/train_final_model.py:/exp_file.py" -e WANDB_API_KEY=<your-api-key> --shm-size=500m train_model
The directory structure of the project looks like this:
├── Makefile <- Makefile with convenience commands like `make data`
├── README.md <- The top-level README for developers using this project.
├── data
│ ├── processed <- The final, canonical data sets for modeling.
│ └── raw <- The original, immutable data dump.
│
├── exp_files <- files to define the experiment configuration
|
├── models <- checkpoint models
│
├── pyproject.toml <- Project configuration file
│
├── requirements.txt <- The requirements file for reproducing the analysis environment
|
├── requirements_dev.txt <- The requirements file for reproducing the analysis environment
│
├── tests <- Test files
│
├── rumexleaves_centernet <- Source code for use in this project.
│
├── submodules <- relevant submodules are stored here
│
└── LICENSE <- MIT LicenseCreated using mlops_template, a cookiecutter template for getting started with Machine Learning Operations (MLOps).
If you find this work useful in your research, please cite:
@article{RumexLeaves-CenterNet,
author = {Güldenring, Ronja and Andersen, Rasmus Eckholdt and Nalpantidis, Lazaros},
title = {Zoom in on the Plant: Fine-grained Analysis of Leaf, Stem and Vein Instances},
journal = {IEEE Robotics and Automation Letters (RA-L)},
year = {2024}
}
Our code is partially based on the following code bases.