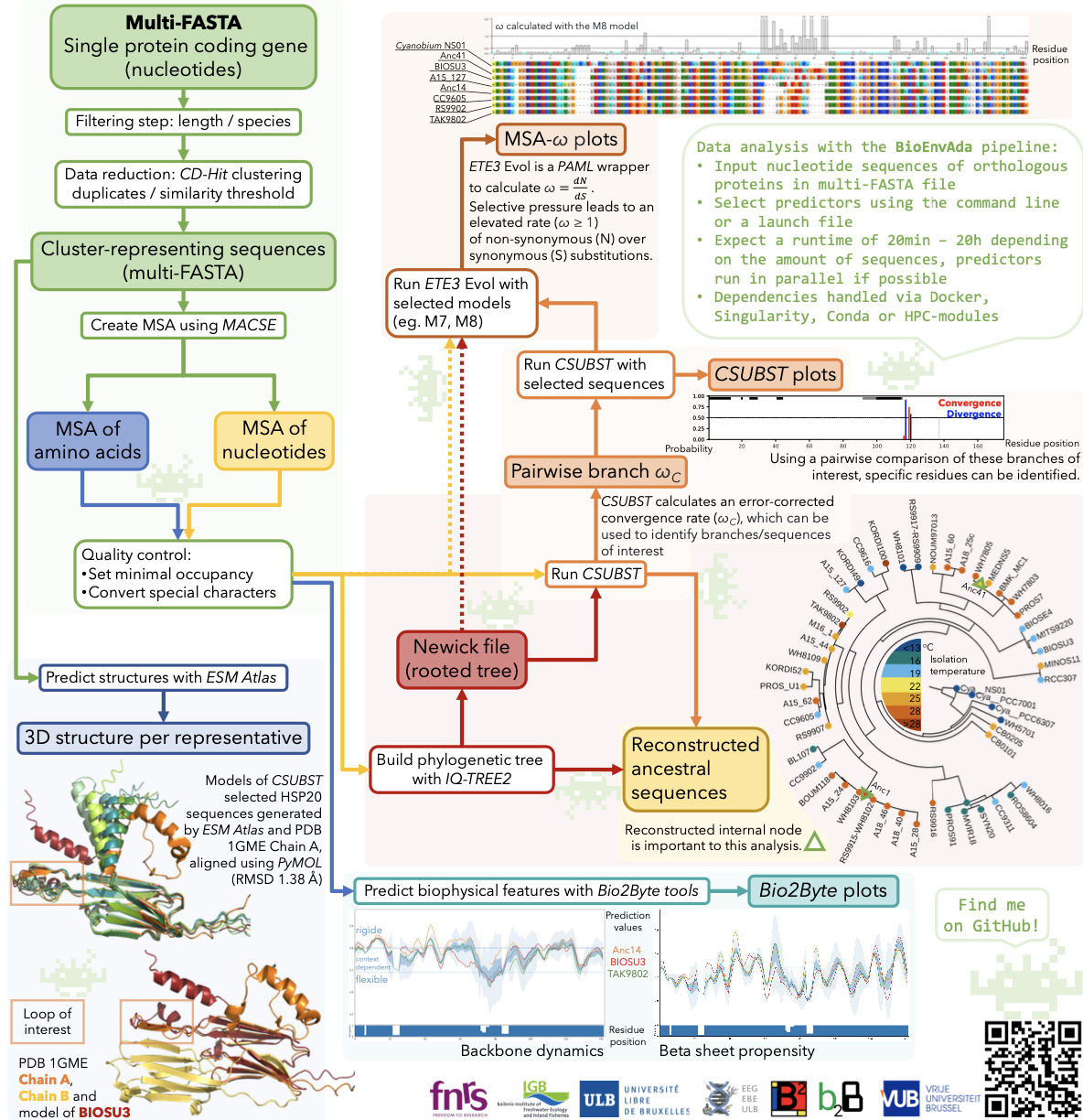
The analysis of protein evolution requires many steps and tools, starting from collecting DNA data to predicting protein structure. We developed a NextFlow (BioEnvAda) pipeline to investigate protein adaptation to changing environmental conditions. It considers multiple aspects of protein evolution comparing changes in amino acid sequences while considering both phylogenetic information and measures of evolutionary pressure. It calculates tendencies for specific biophysical behaviours accounting for the local sequence environments and incorporates predicted 3D structures of a protein.
The default for all parameters in BioEnvAda is false. If you want to use a predictor, add the flag to the command line to turn it on.
Usage in commad line:
nextflow run pipeline.nf
-profile standard,withdocker
--targetSequences ../input_example.fasta
--type 'aa' or 'nuc'
--qc
--clustering 0.85
--relabel
--alignSequences
--efoldmine
--disomine
--agmata
--fetchStructures
--buildTreeEvo \
--outGroup 'Species name to root your tree on'
--csubst
--branchIds '1,2,3'
--eteEvol 'M7,M8'
--selectedProteins 'your,proteins,as,str'
--plotBiophysicalFeatures
--buildLogo
--plotTree
-resume
Alternatively, adapt launch file run_nf.sh
- Cancel a running Nextflow job: Crtl + C
- Pipeline failed to complete:
- to rerun the last job: append -resume to the launch command
- to rerun a specific job: check the
*.nflogfiles last line to get the unique hashnextflow run simsapiper.nf -resume 9ae6b81a-47ba-4a37-a746-cdb3500bee0f
- This can also be used to create plots with different highlighted proteins or different selected branches for csubst, without the need to recalculate all other steps
- Attention: last state will be permanently overwritten
- All intermediate results are unique subdirectory of the directory
work
Find directory hash for each step in*.nflog - Run in the background: launch SIMSApiper in a screen
Hit Crtl + A and Crtl + D to put it in the background
screen -S nextflowalign bash -c ./magic_align.sh
- Input file: --targetSequences path/to/data/file
- Input file type: --type nuc
- NOTE: For input of amino acid sequences use 'aa'
- Set minimal ooccupancy of position in MSA: --qc
- NOTE:
- --qc to remove empty columns in alignment
- --qc 0.85 to set minial occupancy
- Clustering with CD-Hit: --clustering 1
- NOTE:
- --clustering to remove duplicate sequences
- --cluster 0.85 to set similarity cutoff
- Adapt labels to clustering: --relabel
-
Align sequences --alignSequences true
- NOTE:
- –-type aa: residue based MSA with Clustal
- --type nuc: nucleotide based MSA with MACSE
- remove flag to keep pre-aligned file
-
DynaMine : ALWAYS
-
DisoMine : --disomine
-
EFoldMine : --efoldmine
-
AgMata : --agmata
Fetch structures using ESM Atlas (--fetchStructures): false
- Phylo. Tree : --buildTreeEvo
- Species name to root your tree on : --outGroup partialSpeciesID
- Csubst : --csubst
- CsubstSite : --branchIds 1,5
- EteEvol : --eteEvol M7,M8
- Proteins to be highlighted in the plots: --selectedProteins AncNode14,Syn_BIOS_U3
- Plot B2btools : --plotBiophysicalFeatures
- Logo : --buildLogo
- Phylo. Tree plot : --plotTree