This repository contains inference code for Accurate Prediction of Antibody Function and Structure Using Bio-Inspired Antibody Language Model
conda env create -f environment.yml
conda activate BALM
The pre-trained weights of BALM can be downloaded from Google Drive link: pretrained-BALM
from modeling_balm import BALMForMaskedLM
from ba_position_embedding import get_anarci_pos
from transformers import EsmTokenizer
import torch
# an antibody sequence example
input_seq = "AVQLQESGGGLVQAGGSLRLSCTVSARTSSSHDMGWFRQAPGKEREFVAAISWSGGTTNYVDSVKGRFDISKDNAKNAVYLQMNSLKPEDTAVYYCAAKWRPLRYSDNPSNSDYNYWGQGTQVTVSS"
tokenizer = EsmTokenizer.from_pretrained("./tokenizer/vocab.txt", do_lower_case=False, model_max_length=168)
tokenizer_input = tokenizer(input_seq, truncation=True, padding="max_length", return_tensors="pt")
# generate position_ids
tokenizer_input.update(get_anarci_pos(input_seq))
with torch.no_grad():
# please download from Google drive link before
model = BALMForMaskedLM.from_pretrained("./pretrained-BALM/")
# on CPU device
outputs = model(**tokenizer_input, return_dict=True, output_hidden_states=True, output_attentions=True)
# final hidden layer representation [batch_sz * max_length * hidden_size]
final_hidden_layer = outputs.hidden_states[-1]
# final hidden layer sequence representation [batch_sz * hidden_size]
final_seq_embedding = final_hidden_layer[:, 0, :]
# final layer attention map [batch_sz * num_head * max_length * max_length]
final_attention_map = outputs.attentions[-1]
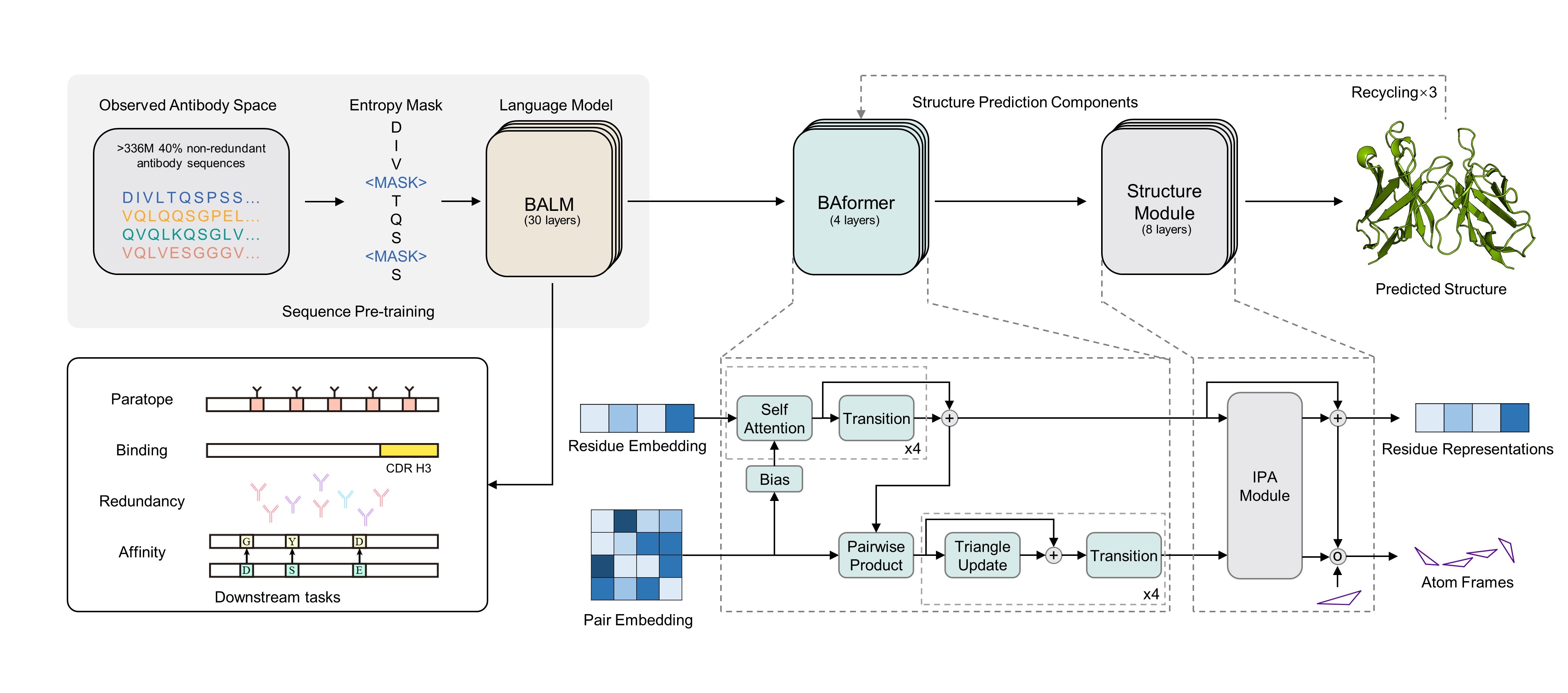
BALMFold is based on BALM to predict antibody tertiary structure with primary sequence. The online server is freely available at BALMFold server. Just try it. :)
If you find our model is useful for you, please cite as:
@article{jing2024accurate,
title={Accurate prediction of antibody function and structure using bio-inspired antibody language model},
author={Jing, Hongtai and Gao, Zhengtao and Xu, Sheng and Shen, Tao and Peng, Zhangzhi and He, Shwai and You, Tao and Ye, Shuang and Lin, Wei and Sun, Siqi},
journal={Briefings in Bioinformatics},
volume={25},
number={4},
pages={bbae245},
year={2024},
publisher={Oxford University Press}
}
The architecture and pre-training process of BALM builds on the ESM and Hugging Face modeling framework. We really appreciate the work of ESM and Hugging Face team.
This source code is licensed under the MIT license found in the LICENSE file in the root directory of this source tree.