A web tool for interactive visualization of the links between taxonomic composition and function in microbiome datasets
Microbiome sequencing studies typically focus on two related but separate questions: 1) which microbial taxa are present, and 2) what genetic functions are present?
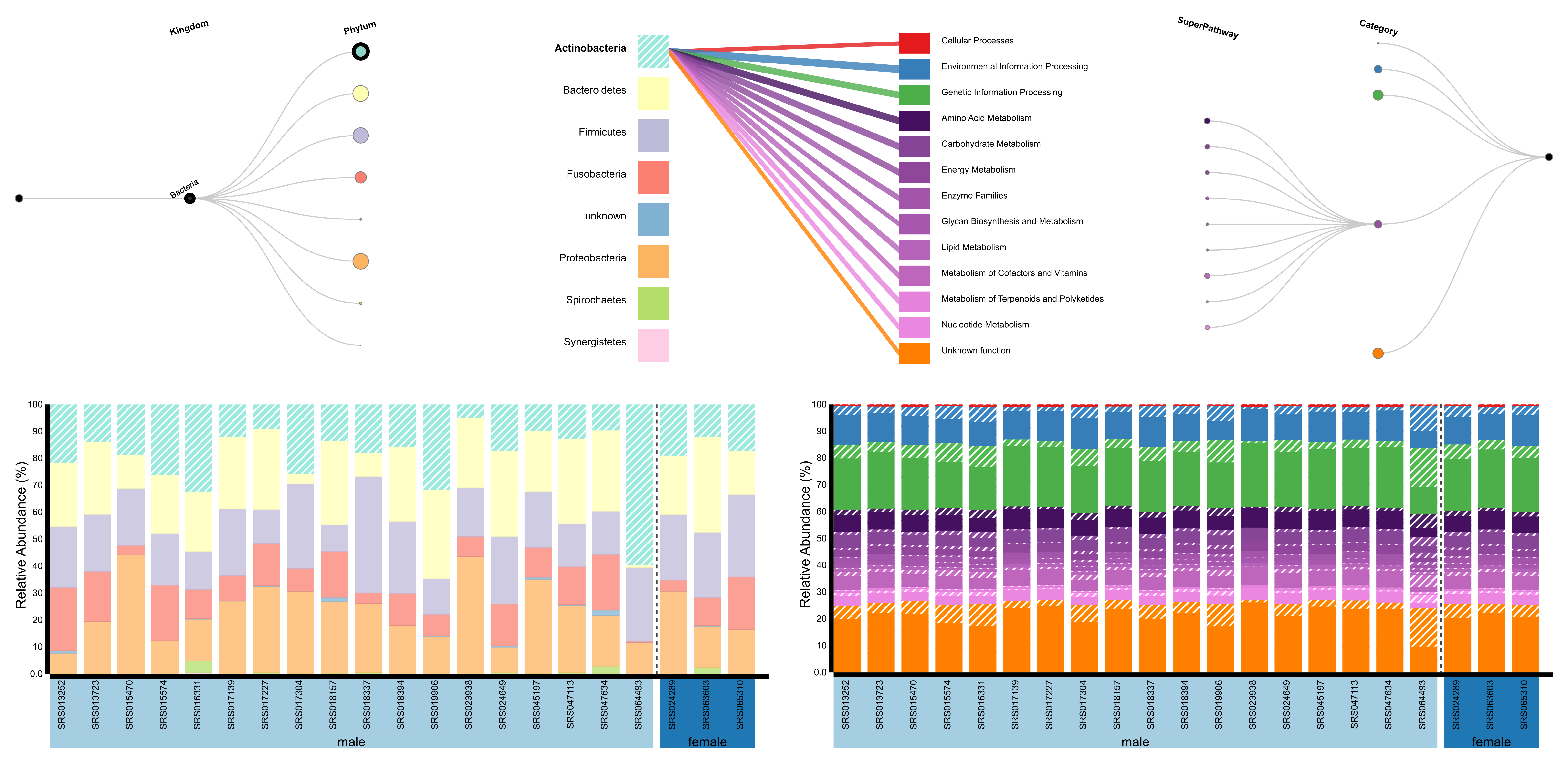
BURRITO is a visualization tool developed to facilitate exploration of both taxonomy and function in microbiome datasets. BURRITO can estimate function abundances from 16S rRNA OTU data, and displays species abundances, function abundances, and the share of each function that can be attributed to each species while simultaneously allowing interactive comparisons between different samples.
BURRITO is located at http://elbo-spice.cs.tau.ac.il/shiny/burrito/
For more information on using BURRITO, please visit the manual at https://borenstein-lab.github.io/burrito/
McNally, C.P., Eng, A., Noecker, C., Gagne-Maynard, W.C., and Borenstein, E. (2018). BURRITO: An Interactive Multi-Omic Tool for Visualizing Taxa–Function Relationships in Microbiome Data. Frontiers in Microbiology 9. doi:10.3389/fmicb.2018.00365