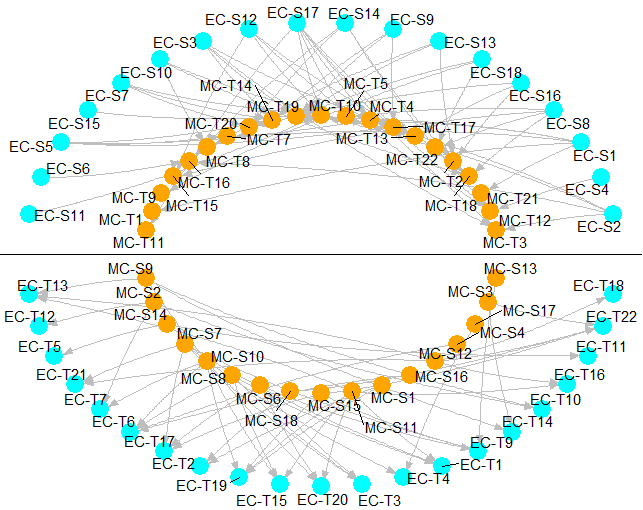
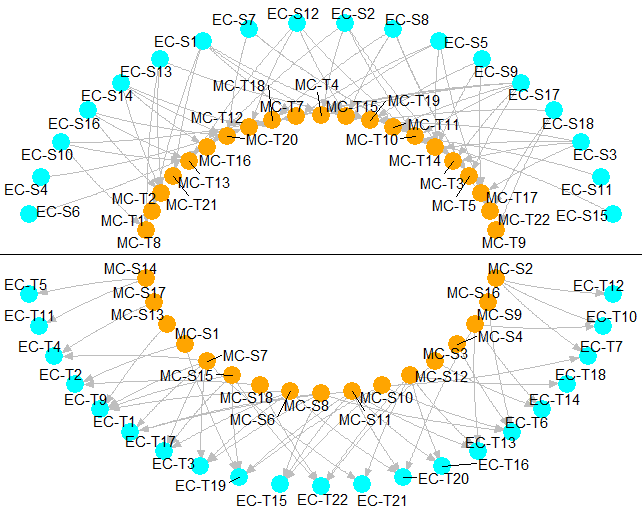
circleInteractionBetween2CellType R function
 =>>
=>>
pcurve princurve:principal curves
ggrepel: Repel overlapping text labels away from each other. https://github.com/slowkow/ggrepel
RSeQC: An RNA-seq Quality Control Package http://rseqc.sourceforge.net
Command Line Interface Creation Kit [:thumbsup:] Python composable command line utility http://click.pocoo.org/
bam2fastx only converts the unmapped reads from the input file, discarding those unmapped reads flagged as QC failed.
bedtools: a powerful toolset for genome arithmetic http://bedtools.readthedocs.io http://www.cureffi.org/2013/11/18/an-mrna-seq-pipeline-using-gsnap-samtools-cufflinks-and-bedtools/
deepTools: tools for exploring deep sequencing data https://deeptools.readthedocs.io/en/develop/
Waddington Plot using ggplot2 R package

- CrossMap
- CrossMap is a program for convenient conversion of genome coordinates (or annotation files) between different assemblies (such as Human hg18 (NCBI36) <> hg19 (GRCh37), Mouse mm9 (MGSCv37) <> mm10 (GRCm38)).
- It supports most commonly used file formats including SAM/BAM, Wiggle/BigWig, BED, GFF/GTF, VCF.
- CrossMap is designed to liftover genome coordinates between assemblies. It’s not a program for aligning sequences to reference genome.
- http://crossmap.sourceforge.net/#convert-gff-gtf-format-files
