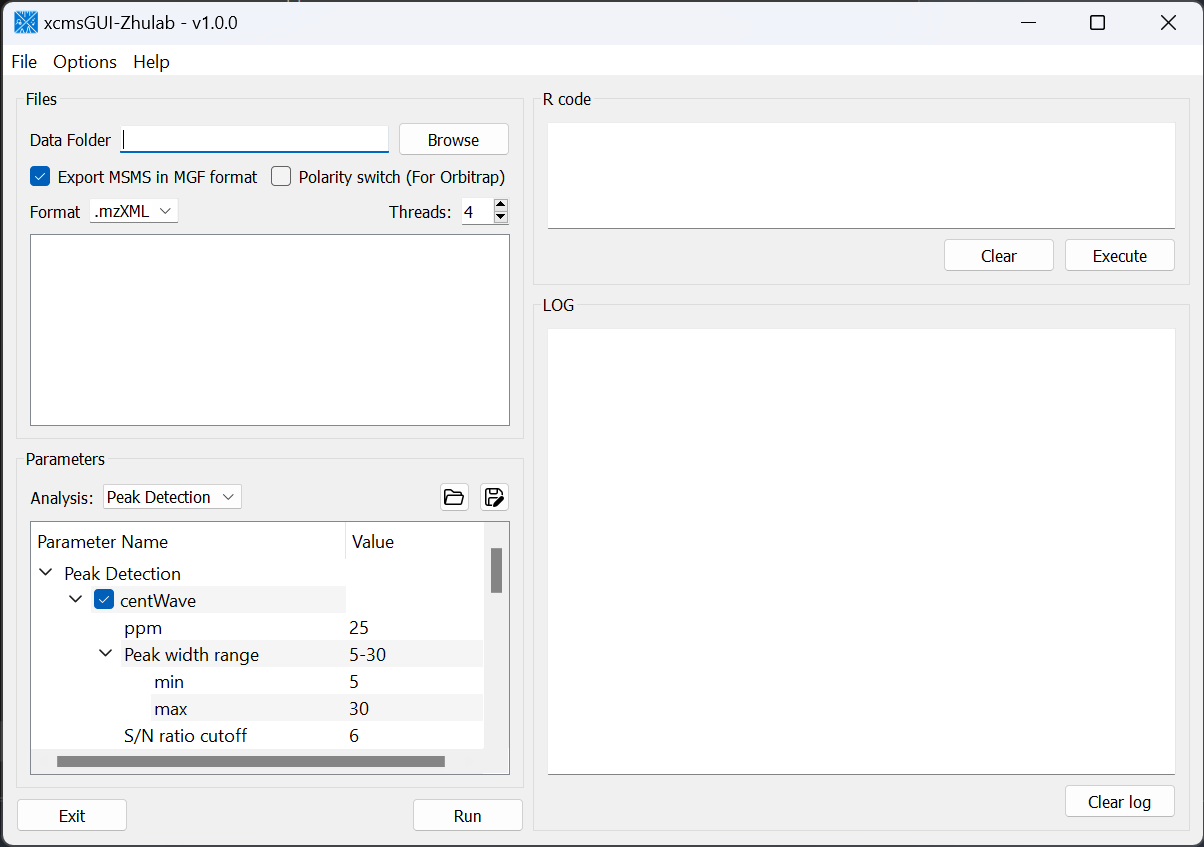
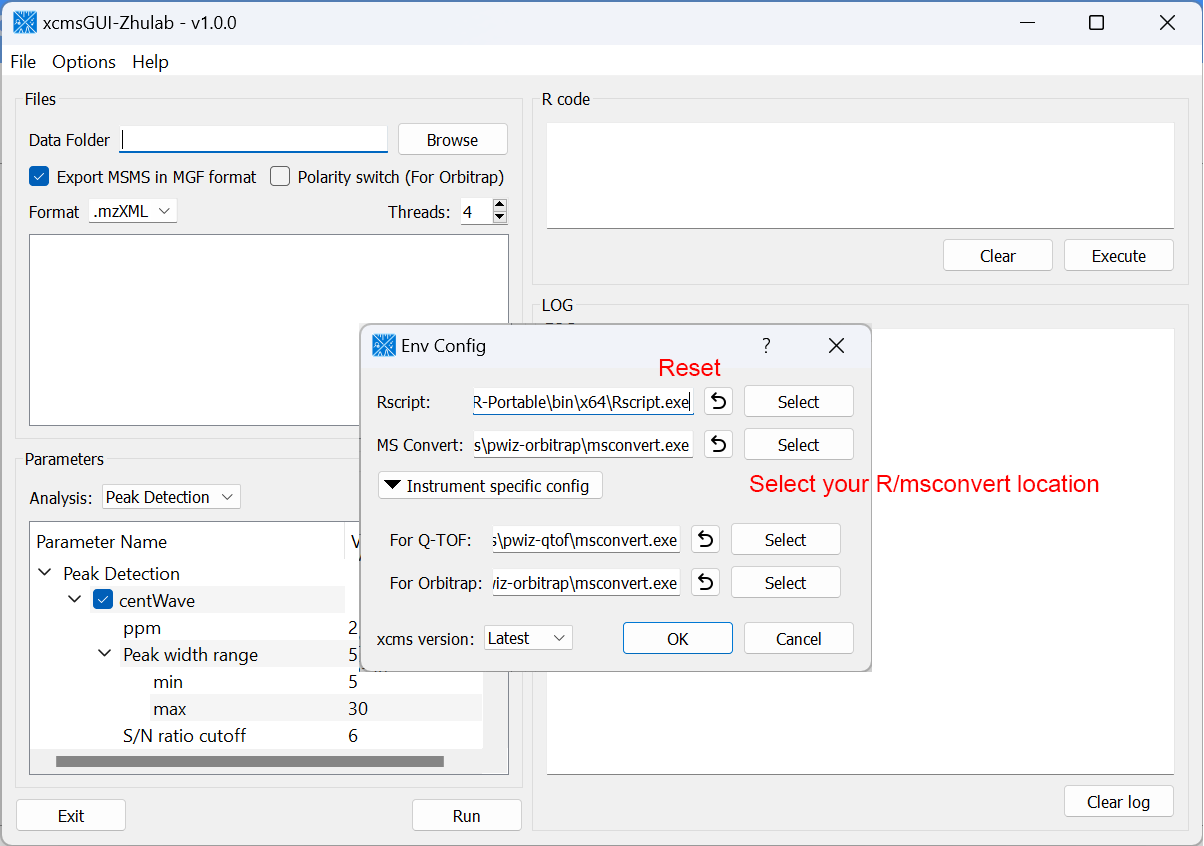
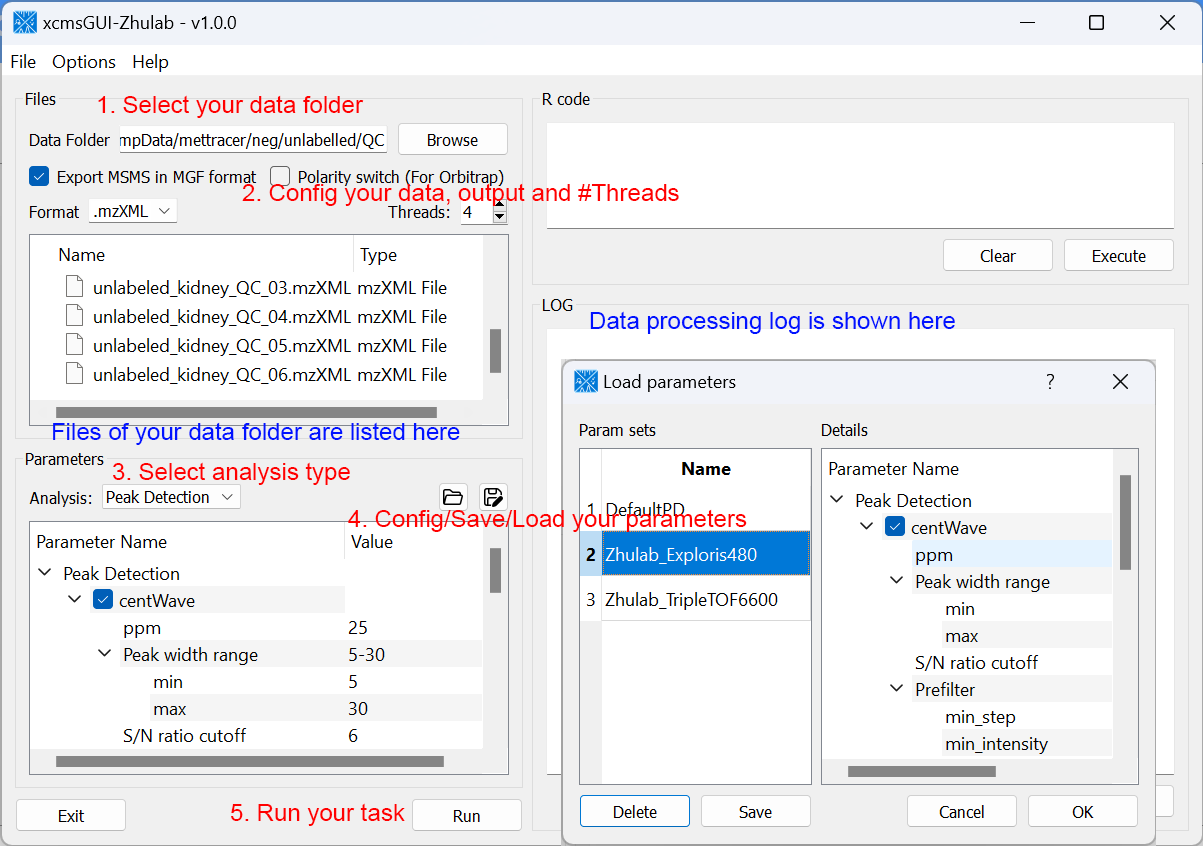
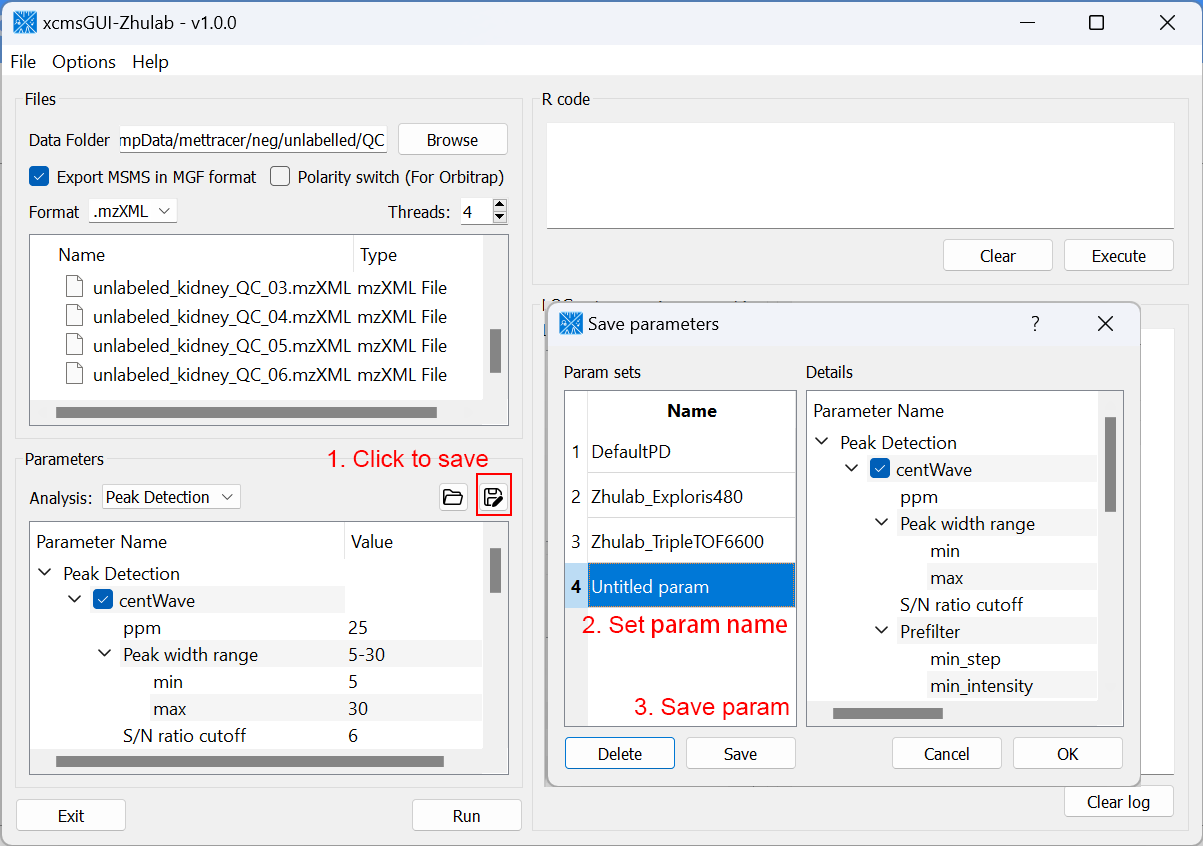
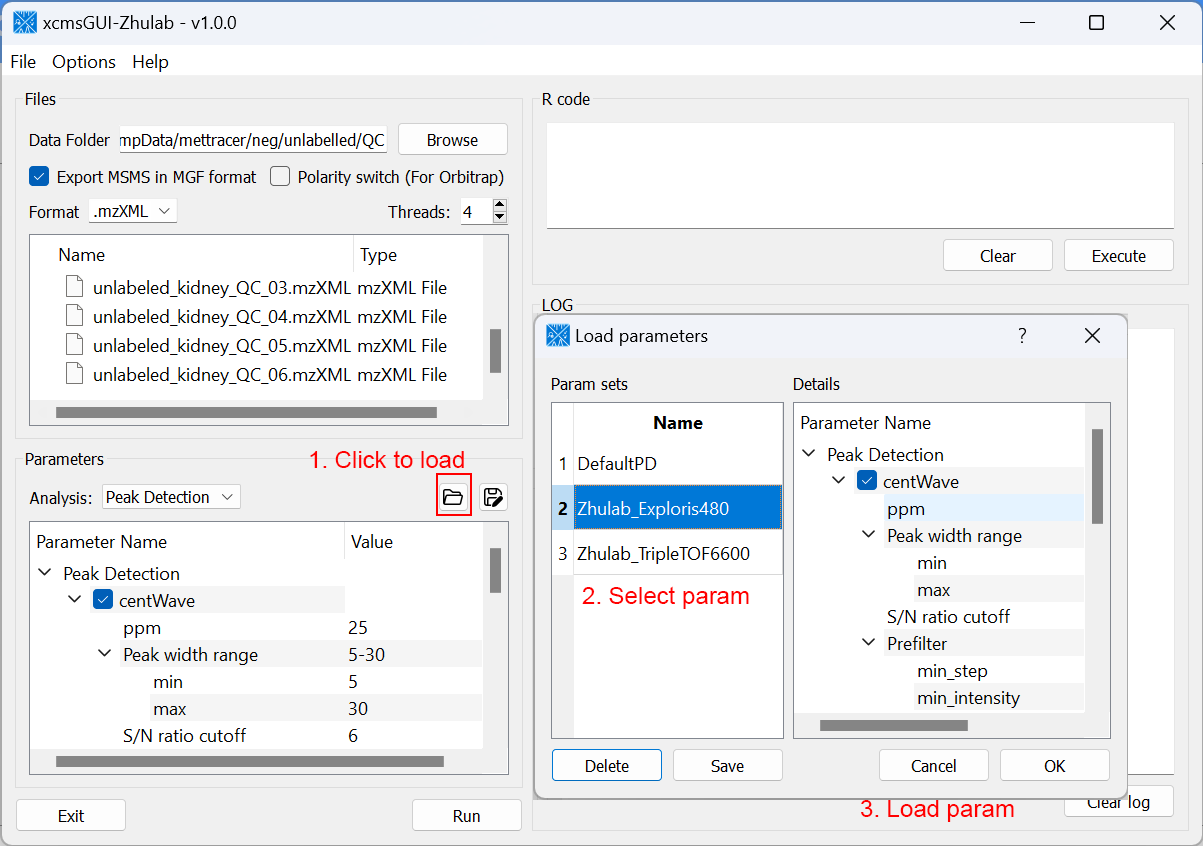
For users' convenience, we developed an all-in-one GUI tool, namely, xcmsGUI-Zhulab, to integrate data conversion (using MSConvert in ProteoWizard) and data pre-processing (using XCMS) in one analysis. Users only need input raw MS data files (.d, .raw, or .wiff) from vendor instruments to xcmsGUI. Then, xcmsGUI enables to perform data conversion and peak detection altogether in one step. Finally, a peak table and MS/MS spectra files (MGF format) are generated as the input files for MetDNA. Sample information file is generated automatically for MetDNA.
Please download the software from release (or just click the following link from GitHub)
- All in one contains
xcmsGUI-ZhulabR-portablePreteoWizard
- To use All in one software, one can just run the xcmsGUI.exe after unzip
xcmsGUI-Zhulab-all.zip
Note
- We support polarity switch mode of ThermoFisher Obitrap instruments (e.g., Exploris 480). Users should just check
Ploarity swith (For Obitrap) and set data format to
and set data format to .raw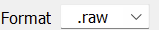
- For MS1 only data, users should uncheck
Export MSMS in MGF Format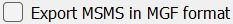
- If only MSMS spectra needed, users should just select
-inAnalysisandRunthe program.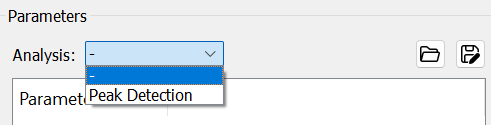
This work is licensed under the Attribution-NonCommercial-NoDerivatives 4.0 International (CC BY-NC-ND 4.0)