Bayesian Categorical Data Analysis
This is a set of tools for Bayesian analysis of categorical data, specifically 2×2 contingency tables.
Use:
beta_binom()for analysis using the Beta-Binomial modelprint(),tidy(),glance(), andplot()to view the resultspresent_bbfit()if you want a nicely formatted table of summaries to include in a presentation or reportupdate()if you have additional data
est_multinom()for estimating multinomial cell probabilities
For more information, see the Tutorial vignette.
Installing
install.packages("devtools")
devtools::install_github("bearloga/BCDA")Usage
All examples will use the following (fake) data:
data <- matrix(c(200, 150, 250, 300), nrow = 2, byrow = TRUE)
colnames(data) <- c('Safe' ,'Dangerous')
rownames(data) <- c('Animals', 'Plants')
(data)| Safe | Dangerous | |
|---|---|---|
| Animals | 200 | 150 |
| Plants | 250 | 300 |
Note that beta_binom() uses the Jeffreys prior by default.
library(BCDA) # options(digits = 2)
set.seed(0)
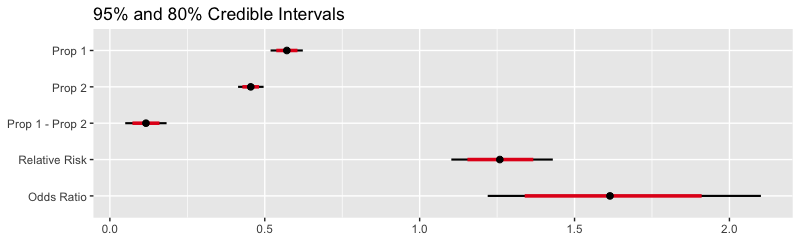
(fit <- beta_binom(data)) term estimate std.error conf.low conf.high
1 p1 0.57 0.027 0.52 0.62
2 p2 0.45 0.021 0.41 0.50
3 prop_diff 0.12 0.034 0.05 0.18
4 relative_risk 1.26 0.083 1.10 1.43
5 odds_ratio 1.61 0.225 1.22 2.10
The credible intervals above are calculated using quantiles. If we have the coda package installed, we can also obtain the high posterior density intervals:
print(fit, interval_type = "HPD") term estimate std.error conf.low conf.high
1 p1 0.57 0.027 0.519 0.62
2 p2 0.45 0.021 0.413 0.50
3 prop_diff 0.12 0.034 0.051 0.18
4 relative_risk 1.26 0.083 1.096 1.42
5 odds_ratio 1.61 0.225 1.198 2.07
plot(fit)Presentation of the results
The package includes a variety of functions for looking at the results from fitting a beta_binom() model. To aid in functional programming, we implemented the tidy() and glance() verbs from David Robinson's broom package for users:
library(magrittr)
fit %>% tidy %>% head(2) term estimate std.error conf.low conf.high
1 p1 0.57 0.027 0.52 0.62
2 p2 0.45 0.021 0.41 0.50
fit %>% glance n1 n2 p1 p2
1 350 550 57.12% (51.88%, 62.29%) 45.48% (41.38%, 49.63%)
This is perfectly okay in an interative data analysis scenario, but not when presenting the results in a report. glance() is actually a special case of the present_bbfit() function which generates all those nicely formatted credible intervals but outputs a Markdown/LaTeX-formatted table by default:
present_bbfit(fit)| Group 1 | Group 2 | Pr(Success) in Group 1 | Pr(Success) in Group 2 | Difference | Relative Risk | Odds Ratio |
|---|---|---|---|---|---|---|
| 350 | 550 | 57.12% (51.88%, 62.29%) | 45.48% (41.38%, 49.63%) | 11.64% (4.96%, 18.30%) | 1.26 (1.10, 1.43) | 1.61 (1.22, 2.10) |
The point estimates include credible intervals by default but these can be turned off:
present_bbfit(fit, conf_interval = FALSE, digits = 3)| Group 1 | Group 2 | Pr(Success) in Group 1 | Pr(Success) in Group 2 | Difference | Relative Risk | Odds Ratio |
|---|---|---|---|---|---|---|
| 350 | 550 | 57.122% | 45.479% | 11.643% | 1.259 | 1.614 |
Since the underlying code uses tidy() to compute the summaries, we can specify a particular credible level and the type of interval we want (e.g. highest posterior density):
present_bbfit(fit, conf_level = 0.8, interval_type = "HPD")| Group 1 | Group 2 | Pr(Success) in Group 1 | Pr(Success) in Group 2 | Difference | Relative Risk | Odds Ratio |
|---|---|---|---|---|---|---|
| 350 | 550 | 57.12% (53.68%, 60.50%) | 45.48% (42.74%, 48.10%) | 11.64% (7.22%, 15.92%) | 1.26 (1.15, 1.36) | 1.61 (1.30, 1.87) |
It also supports multiple models, which can be provided as a named or an unnamed list. See the example below.
Updating the posterior
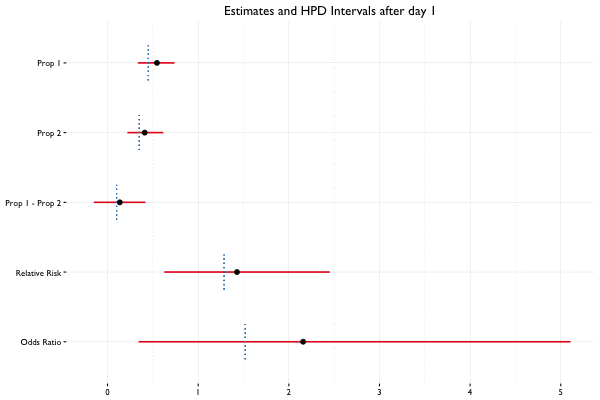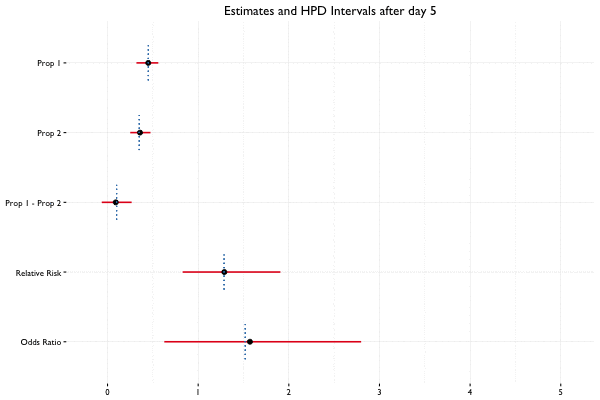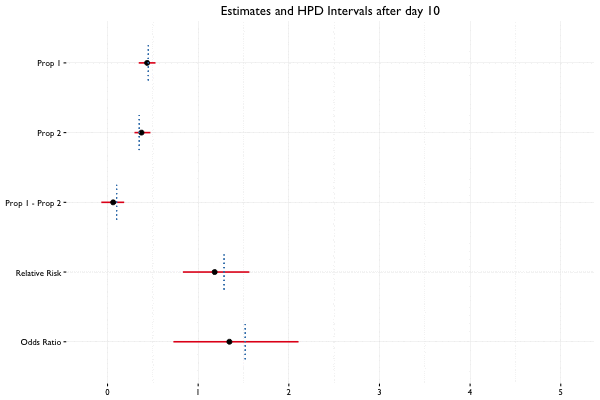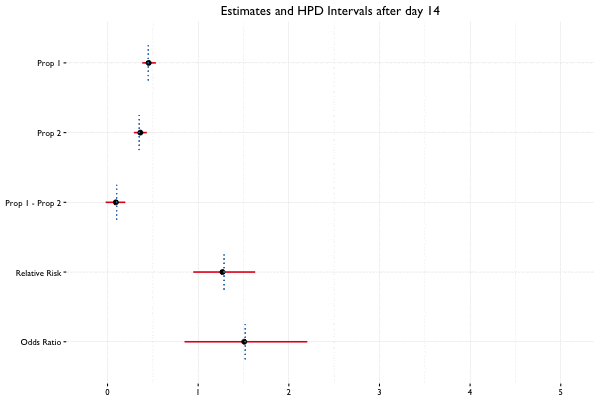
In Bayesian statistics, we can reuse a previously computed posterior as a prior if we have additional data, allowing us to update the parameter estimates as new data becomes available. Suppose we collect 40 observations from 2 groups (20 per group) on the first day of the A/B test, and 10 observations per day for the next 2 weeks. Here we see what happens when we update the posterior with additional data on a daily basis:
Example Code
fit_2 <- update(fit, x = c(100, 200), n = c(400, 600))
present_bbfit(list("Day 1" = fit, "Day 2" = fit_2))| Group 1 | Group 2 | Pr(Success) in Group 1 | Pr(Success) in Group 2 | Difference | Relative Risk | Odds Ratio | |
|---|---|---|---|---|---|---|---|
| Day 1 | 350 | 550 | 57.12% (51.88%, 62.29%) | 45.48% (41.38%, 49.63%) | 11.64% (4.96%, 18.30%) | 1.26 (1.10, 1.43) | 1.61 (1.22, 2.10) |
| Day 2 | 750 | 1150 | 39.96% (36.45%, 43.24%) | 39.18% (36.23%, 42.01%) | 0.78% (-3.73%, 5.08%) | 1.02 (0.91, 1.13) | 1.04 (0.85, 1.24) |
If you purrrfer, you can achieve similar results the following way:
list("1" = fit, "2" = fit_2) %>%
purrr::map_df(present_bbfit, raw = TRUE, .id = "Day") %>%
# ...other manipulations... %>%
knitr::kable()| Day | Group 1 | Group 2 | Pr(Success) in Group 1 | Pr(Success) in Group 2 | Difference | Relative Risk | Odds Ratio |
|---|---|---|---|---|---|---|---|
| 1 | 350 | 550 | 57.12% (51.88%, 62.29%) | 45.48% (41.38%, 49.63%) | 11.64% (4.96%, 18.30%) | 1.26 (1.10, 1.43) | 1.61 (1.22, 2.10) |
| 2 | 750 | 1150 | 39.96% (36.45%, 43.24%) | 39.18% (36.23%, 42.01%) | 0.78% (-3.73%, 5.08%) | 1.02 (0.91, 1.13) | 1.04 (0.85, 1.24) |
See also
Other packages for Bayesian analysis of A/B tests include: LearnBayes (GPL), conting (GPL), bandit (GPL), testr (MIT).
Please note that this project is released with a Contributor Code of Conduct. By participating in this project you agree to abide by its terms.