PYthon Neural Analysis Package.
pynapple is a light-weight python library for neurophysiological data analysis. The goal is to offer a versatile set of tools to study typical data in the field, i.e. time series (spike times, behavioral events, etc.) and time intervals (trials, brain states, etc.). It also provides users with generic functions for neuroscience such as tuning curves and cross-correlograms.
- Free software: GNU General Public License v3
- Documentation: https://peyrachelab.github.io/pynapple
- Notebooks and tutorials : https://peyrachelab.github.io/pynapple/notebooks/pynapple-quick-start/
- Collaborative repository: https://github.com/PeyracheLab/pynacollada
Note 📃 If you are using pynapple, please cite the following biorxiv paper
The best way to install pynapple is with pip within a new conda environment :
$ conda create --name pynapple pip python=3.8
$ conda activate pynapple
$ pip install pynapple
or directly from the source code:
$ conda create --name pynapple pip python=3.8
$ conda activate pynapple
$ # clone the repository
$ git clone https://github.com/PeyracheLab/pynapple.git
$ cd pynapple
$ # Install in editable mode with `-e` or, equivalently, `--editable`
$ pip install -e .
Note The package is now using a pyproject.toml file for installation and dependencies management. If you want to run the tests, use pip install -e .[dev]
This procedure will install all the dependencies including
- pandas
- numpy
- scipy
- numba
- pynwb 2.0
- tabulate
- pyqt5
- pyqtgraph
- h5py
For spyder users, it is recommended to install spyder after installing pynapple with :
$ conda create --name pynapple pip python=3.8
$ conda activate pynapple
$ pip install pynapple
$ pip install spyder
$ spyder
Warning note for Windows users: on a multi-user Windows, make sure you open the conda prompt with administrative access:
run as administrator; otherwise directory paths for some dependencies may be missing from the PYTHONPATH environment variable. The most common is the error in importing PyQt5. In case of such errors, right click on your conda prompt and selectrun as administrator, activate your pynapple environment, and install the said package again (e.g. pip install PyQt) so that the paths are properly saved by Windows.
Warning note for Mac users with M1/M2 chips: PyQt5 may fail to install. In the Application folder, make a copy of the Terminal app (or any other console you're using), and call the second one, for exampe, 'Terminal-Rosetta'. Right click->Get Info and click on 'Open using Rosetta'. Open this terminal and enter the following instructions (you need to install virtualenv before)
$ /usr/bin/python3 -m venv pynapple
$ (pynapple) pip install --upgrade pip
$ (pynapple) pip install pynapple
Open python and try:
import pynapple as nap
nap.load_session()The data loader should open in a new window.
After installation, you can now import the package:
$ python
>>> import pynapple as nap
You'll find an example of the package below. Click here to download the example dataset. The folder includes a NWB file containing the data (See this notebook for more information on the creation of the NWB file).
import numpy as np
import pandas as pd
import pynapple as nap
from matplotlib.pyplot import *
data_directory = '/your/path/to/A2929-200711'
# LOADING DATA
data = nap.load_session(data_directory, 'neurosuite')
spikes = data.spikes
position = data.position
wake_ep = data.epochs['wake']
# COMPUTING TUNING CURVES
tuning_curves = nap.compute_1d_tuning_curves(group = spikes,
feature = position['ry'],
ep = position['ry'].time_support,
nb_bins = 120,
minmax=(0, 2*np.pi) )
# PLOT
figure()
for i in spikes:
subplot(6,7,i+1, projection = 'polar')
plot(tuning_curves[i])
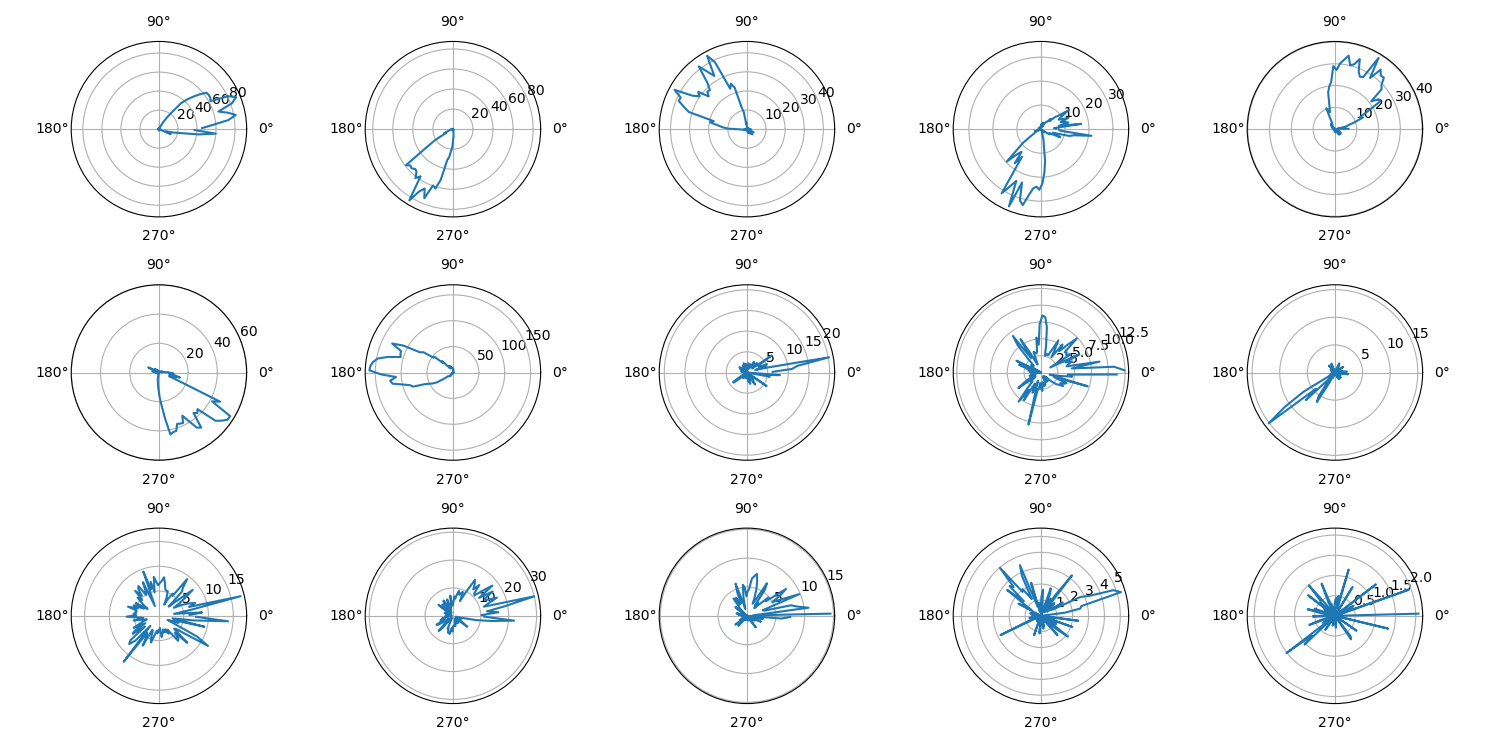
show()Shown below, the final figure from the example code displays the firing rate of 15 neurons as a function of the direction of the head of the animal in the horizontal plane.
Special thanks to Francesco P. Battaglia (https://github.com/fpbattaglia) for the development of the original TSToolbox (https://github.com/PeyracheLab/TStoolbox) and neuroseries (https://github.com/NeuroNetMem/neuroseries) packages, the latter constituting the core of pynapple.
This package was developped by Guillaume Viejo (https://github.com/gviejo) and other members of the Peyrache Lab.
Logo: Sofia Skromne Carrasco, 2021.