YouTube Video: https://youtu.be/J8wMq6xQ-Lc
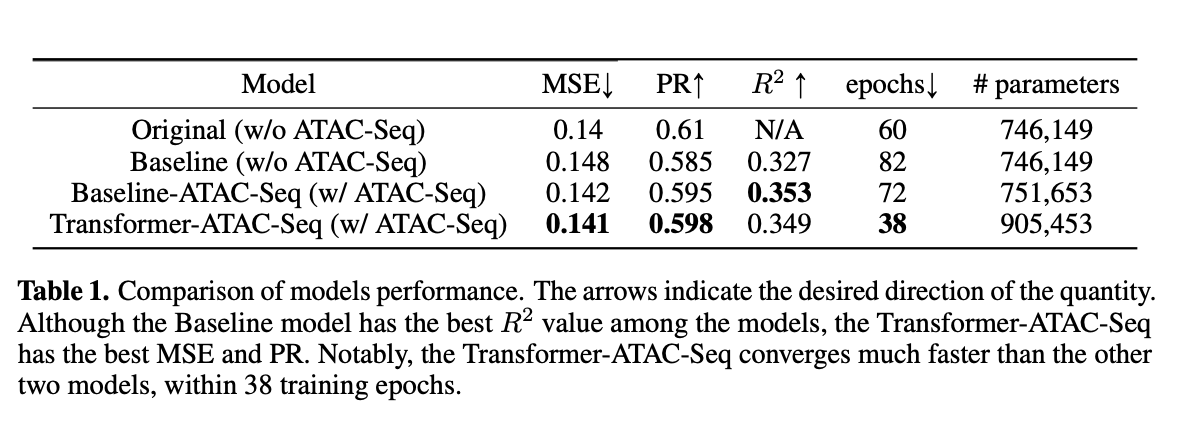
In this project, we reproduced the paper, titled “Akita, a CNN model for predicting 3D genome folding from DNA sequence.” and re-implemented the Akita Network, which is a convolutional neural network (CNN) that transforms input DNA sequence into predicted locus-specific genome folding. We then tried to improve the model performance in two ways: (1) leverage ATAC-Seq data, and (2) add Transformer encoder layers. Same as the original paper, we tested the performance on the held-out test dataset.
We firstly construct the Akita network and test it on the held-out test dataset. The hyper-parameters are the same as reported in the paper. Small tweaks: replace residual CNN blocks with "pre-activation" residual CNN blocks.
ATAC-Seq data was concatenated with corresponding DNA sequences. All the other settings are the same as experiment 1.
Four additional Transformer encoder layers were added into the Akita network to help capture the long-range relationship. The encoder layers are implemented as the "pre-layernorm" variant to stablize training.
- The jupyter notebook './project.ipynb' contains the three experiments decribed above.
- The script './custom_models.py' contains our own implementations of the models, modules, and helper functions used in the project.
- The directory './Datasets' contains all the datasets used in the project. './Datasets/get_data.sh' are used to get train, validation, test DNA sequences and Hi-C maps. './Datasets/Sample_0902.bed' contains ATAC-Seq reads.
- The directory './Figures' contains related figures such as training processes, model architecture inllustrations etc.
- The directory './checkpoints' contains trained weights.
- Python 3.8.13
- Tensorflow 2.8.0
- Basenji
.
├── Datasets
│ ├── Sample_0902.bed
│ └── get_data.sh
├── Figures
│ ├── MHA.pdf
│ ├── Residual_refine_1D:2D.pdf
│ ├── Transformer_encoder.pdf
│ ├── akita_architecture.png
│ ├── baseline_1.png
│ ├── baseline_2.png
│ ├── baseline_3.png
│ ├── model_overall.pdf
│ ├── orig_1.png
│ ├── orig_2.png
│ ├── orig_3.png
│ ├── re_imp_1.png
│ ├── re_imp_2.png
│ ├── re_imp_3.png
│ ├── residual_downsample_1d.pdf
│ ├── result.png
│ ├── transf_1.png
│ ├── transf_2.png
│ └── transf_3.png
├── README.md
├── checkpoints
│ ├── 20220501-121443.data-00000-of-00001
│ ├── 20220501-121443.index
│ ├── 20220501-211604.data-00000-of-00001
│ ├── 20220501-211604.index
│ ├── 20220502-090312.data-00000-of-00001
│ └── 20220502-090312.index
├── custom_models.py
└── project.ipynb
3 directories, 30 files