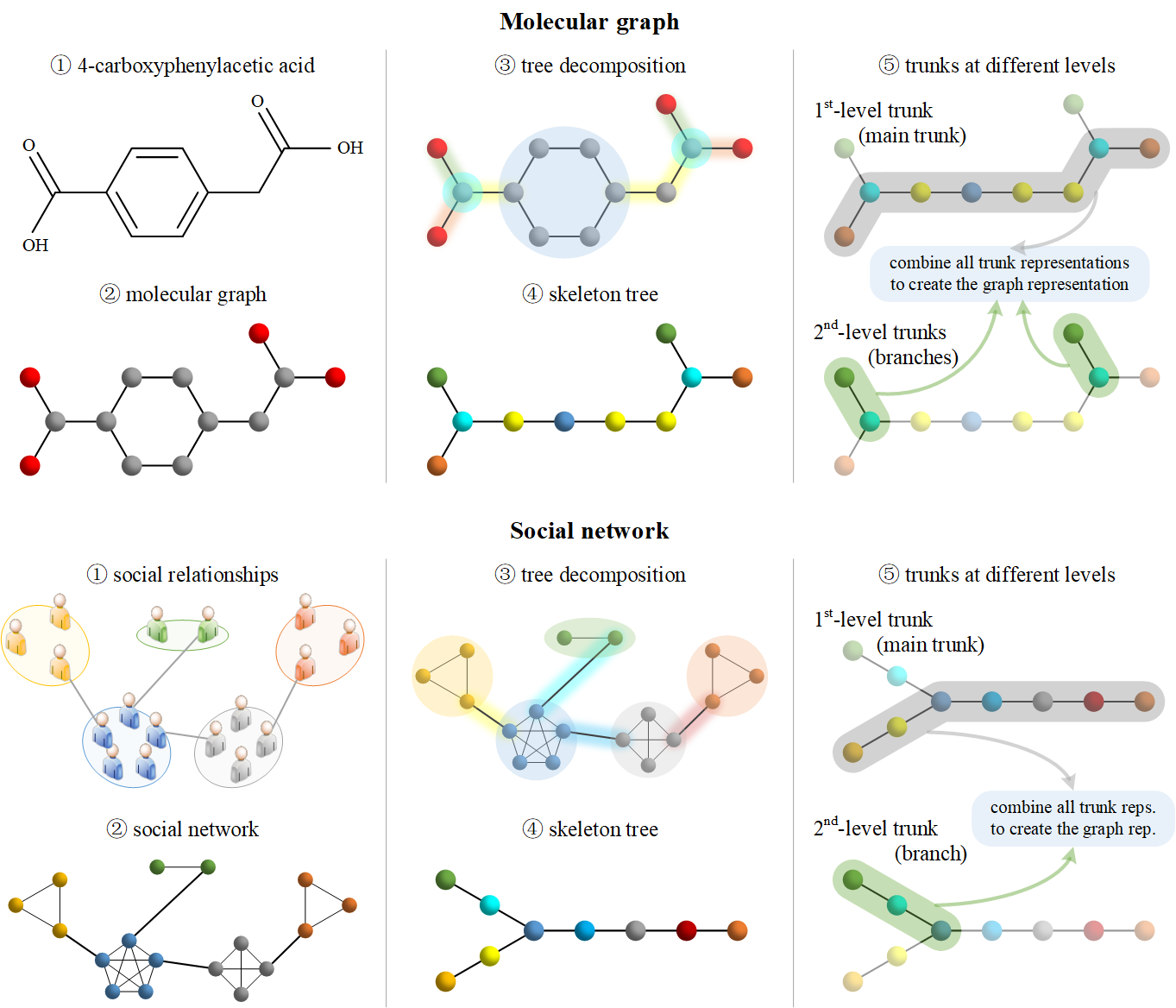
The official implementation of Growing Like a Tree: Finding Trunks From Graph Skeleton Trees (TPAMI 2023).
Follow the steps below to set up the virtual environment.
Create and activate the environment:
conda create -n gtr python=3.6
conda activate gtrInstall dependencies in the listed order:
pip install rdkit-pypi==2021.3.5
pip install -r requirements.txt
pip install torch==1.4.0+cu100 -f https://download.pytorch.org/whl/torch_stable.html
pip install torch_scatter==2.0.4 -f https://pytorch-geometric.com/whl/torch-1.4.0+cu100.html
pip install torch_sparse==0.6.1 -f https://pytorch-geometric.com/whl/torch-1.4.0+cu100.html
pip install torch-geometric==1.7.0
pip install ogb==1.2.6Reproduce the results reported in Table 3. Repeat the experiment 100 times using the same evaluation protocol as Karhadkar et al. to obtain the reported results.
python main_tu.py --name MUTAG --batch_size 16 --hidden_dim 64 --num_layers 4 --dropout 0.5 --lr 0.001 --lr_factor 0.5 --lr_limit 1e-5 --max_level 5python main_tu.py --name ENZYMES --batch_size 16 --hidden_dim 64 --num_layers 4 --dropout 0.5 --bidirectional --lr 0.001 --lr_factor 0.5 --lr_limit 1e-5 --max_level 10python main_tu.py --name PROTEINS --batch_size 16 --hidden_dim 64 --num_layers 4 --dropout 0.5 --bidirectional --lr 0.001 --lr_factor 0.1 --lr_limit 5e-6 --max_level 10python main_tu.py --name IMDB-BINARY --batch_size 16 --hidden_dim 64 --num_layers 4 --dropout 0.5 --lr 0.001 --lr_factor 0.5 --lr_limit 2e-5 --max_level 2python main_tu.py --name COLLAB --batch_size 32 --hidden_dim 64 --num_layers 4 --dropout 0.5 --lr 0.001 --lr_factor 0.5 --lr_limit 2e-5 --max_level 12Reproduce the results reported in Table 4.
Repeat the experiment 10 times using the same evaluation protocol as Hu et al. to obtain the reported results.
python main_ogbhiv.py --name ogbg-molhiv --batch_size 128 --hidden_dim 128 --num_layers 4 --dropout 0.5 --lr 0.0003 --lr_factor 0.5 --lr_limit 5e-5 --max_level 4Repeat the experiment 4 times using the same evaluation protocol as Dwivedi et al. to obtain the reported results.
python main_pepfunc.py --name Peptides-func --batch_size 128 --hidden_dim 256 --num_layers 4 --dropout 0.5 --lr 0.0004 --lr_factor 0.5 --lr_limit 2e-5 --max_level 12Repeat the experiment 4 times using the same evaluation protocol as Dwivedi et al. to obtain the reported results.
python main_pepstrc.py --name Peptides-struct --batch_size 128 --hidden_dim 256 --num_layers 3 --dropout 0.2 --lr 0.0004 --lr_factor 0.5 --lr_limit 2e-5 --max_level 12We provide the IPython Notebook file visualization.ipynb for reproducing the visualizations shown in Figure 3 (Section 5.6).
If you find this code or our GTR paper helpful for your research, please cite our paper using the following information:
@article{huang2023growing,
title = {Growing Like a Tree: Finding Trunks From Graph Skeleton Trees},
author = {Huang, Zhongyu and Wang, Yingheng and Li, Chaozhuo and He, Huiguang},
journal = {IEEE Transactions on Pattern Analysis and Machine Intelligence},
year = {2023}
}