OpenMat is a parallel sparse matrix data generator based on PDE discretization designed for intelligent iterative methods. It can not only generate matrix data, but also compute the label of the matrix. In OpenMat, both matrix generation and label computation can be accelerated through task parallelism.
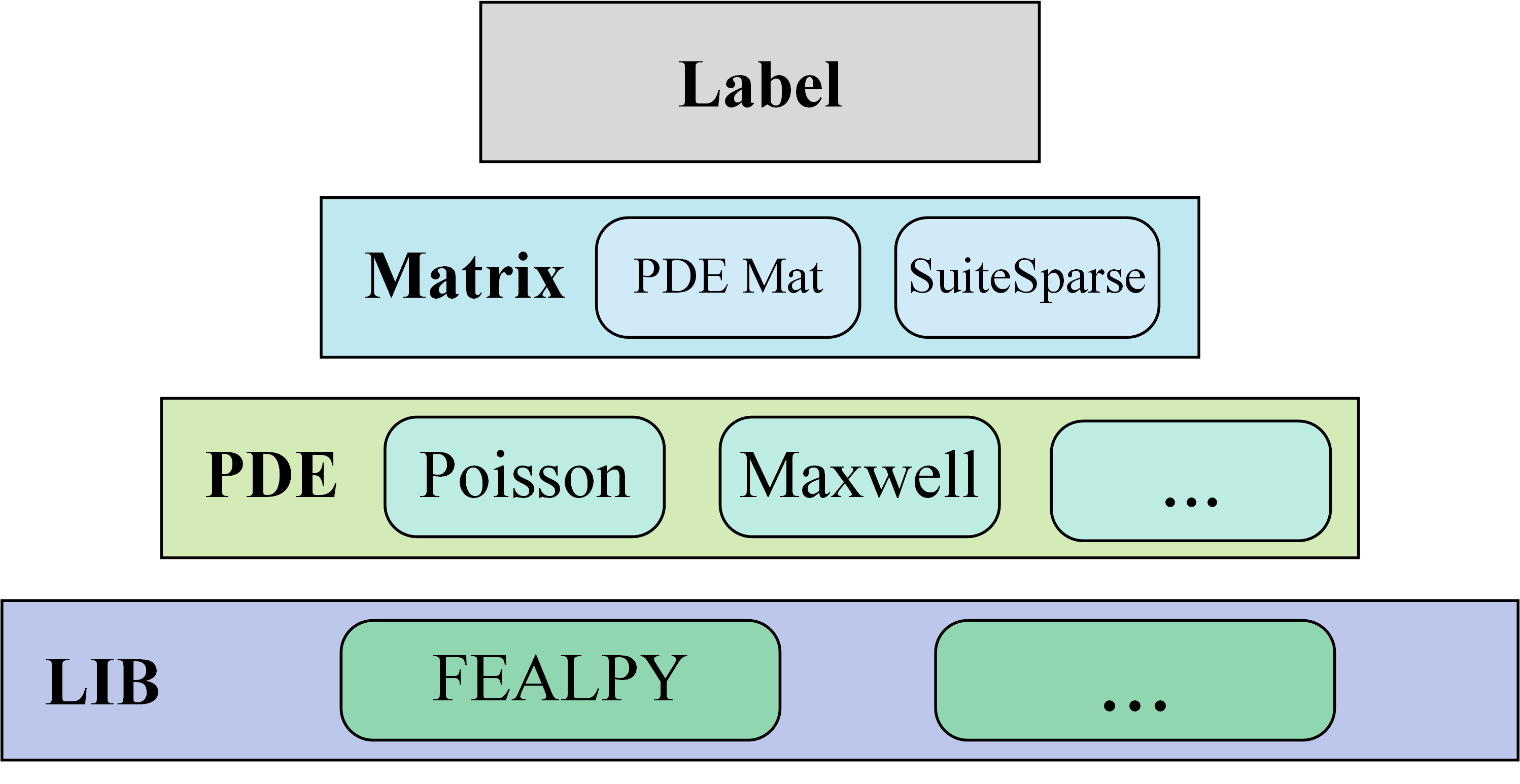
The architecture of OpenMat is:
Based on FEALPY, OpenMat can generate matrices with different sizes and properties by adjusting discretization parameters. In the future, we will add more PDEs, and support other libraries, such as FEniCS, MFEM. The features of OpenMat include:
- Label Computation: In addition to matrix generation,
OpenMatutilizes various iterative methods provided byPETScto solve the the same linear equation, and the fastest iterative method is the label of the matrix. - Support for Multiple Programming Languages: Both matrix generation and label computation can be implemented by different programming languages (
C/C++,python, etc. ) - Support for SuiteSparse: Providing the matrix ID is sufficient to download the corresponding matrix in SuiteSparse matrix collection
- Parallel Computing: Multi-task parallelism
- Checkpoint Restart: The execution can be interrupted at any point during runtime. The program would skip previously completed computations and resume from the last checkpoint, avoiding starting from scratch.
- Matrix Reconstruction Based on Configuration File:
OpenMatemploys theGenerateMat()function to create a matrix and simultaneously saves the input parameters to the correspondingJSONfile. Therefore, by providing the matrix generation program and the correspondingJSONfile,OpenMatcan regenerate the matrix
OpenMat:git clone git@github.com:zhf-0/matrix.gitor download the tar file directly- Matrix generation and label computation can be used independently which need different softwares
- Matrix generation: install
FEALPY(installation) andssgetpy(download matrices from SuiteSpare Matrix Collection, installation) - Label computation: install
petscandpetsc4py(offical PETSc doc)
- Matrix generation: install
Note that: OpenMat can be used in cluster and desktop envirorments. For now, when it is used in the desktop, the mpi must be openmpi because of the processes binding
Using Poisson equation as the example
# the dir that stores all json files
json_dir = './JsonFiles/'
# the dir that stores all generated matrces
mat_dir = './MatData/'
# the name of the bash script that store all commands
script_path = gen.sh
# the header of the script `gen.sh`
header = ['#!/bin/bash \n']
# the footer of the script `gen.sh`
footer = ['echo finished !! \n']
# define a empty list to store all commands, then write to `script_path`
contents = []
# index of the matrix, use 100 as an example
idx = 100
# the type of output matrix, SciCSR: scipy.sparse.csr_matrix(default)
# SciCOO: scipy.sparse.coo_matrix
# COO: coo in txt format
mat_type = 'SciCSR'
# matrix name, the name has to be choosen from the following list:
# if the format is 'SciCSR', the name is 'scipy_csr{index}.npz'
# if the format is 'SciCOO', the name is 'scipy_coo{index}.npz'
# if the format is 'COO', the name is 'coo{index}.npz'
mat_path = os.path.join(mat_dir,f'scipy_csr{idx}.npz')
# output the right hand side vector
need_rhs = 1
# don't need to output the right hand side vector
# need_rhs = 0
# pre-defined parameters in Poisson equation, including
# nx (mesh size in x axis), ny, etc
para = pde1.Para()
# define new parameters
para.DefineFixPara('mat_type',mat_type)
para.DefineFixPara('mat_path',mat_path)
para.DefineFixPara('need_rhs',need_rhs)
# create a json file that save all parameters of the generation
json_path = os.path.join(json_dir,f'result{i}.json')
json_result = {}
#the index of poisson problem is 1 in OpenMat
json_result['PDE_idx'] = 1
# parameter.para is the dict that includes all values of the parameter
# copy all paremeters to the json file
json_result['GenMat'] = para.para
# save json file
with open(json_path,'w',encoding='utf-8') as f:
json.dump(json_result,f,indent=4)
# create the command with command line parameters
# such as `line = --nx 10, --ny 10 --mat_type tri ...`
line = ' '
for k,v in para.para.items():
line = line + f' --{k} {v} '
one_command = 'python ./PDEs/poisson_lfem_mixedbc_2d.py' + line + '\n'
# add the command into the list
contents.append(one_command)
# the codes above only concern about one matrix
# if the commands for all matrices is generated, then save them to file
with open(script_path,'w',encoding='utf-8') as f:
f.writelines(header)
f.writelines(contents)
f.writelines(footer)More details can be found in main_generate.py.
using petsc
# the dir that stores all yaml files, each yaml file store the
# solving result of the matrix only temporarily.
yaml_dir = './YamlFiles'
# the same dir as in the matrix generation step,
# the program will extract the information from yaml file
# to the json file
json_dir = './JsonFiles'
# change the matrix file in `mat_dir` in matrix generation step
# to petsc binary file and save the binary file into another dir
mat_dir = 'PetscMat'
# each iterative method will solve 3 times for average
batch_size = 3
# the number of scripts that will be generated
num_task = 8
# the command used to sovle linear equations based on petsc
command = './Solve/rs -ksp_type {} -pc_type {} -mat_file {} -yaml_file {} \n'
# all matrix indexs, use [0,1,...999] as an example
idx_list = list(range(1000))
# the name template of the matrix file generated in last step
mat_template = './MatData/scipy_csr{}.npz'
# the name of the bash script that store all commands
script_file = 'solve.sh'
# the header of the script
header = ['#!/bin/bash \n']
# the footer of the script
footer = ['echo finished !! \n']
# create an instance of the class and begin to process
a = ParTaskParRunCluster(json_dir,yaml_dir,mat_dir,batch_size,idx_list,num_task)
a.Process(mat_template)
a.GenerateScript(script_file,header,footer,command)