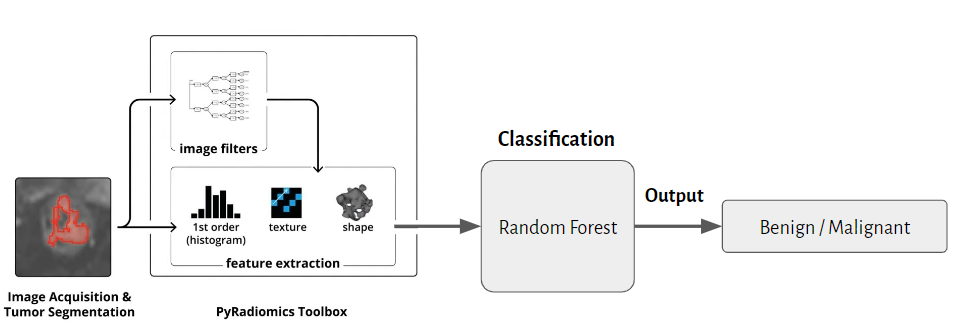
We used CT images and masks of renal tumors to extract radiomics features, and then used a random forest classification model to classify tumors as benign or malignant.
CT image
MaskThe picture source: https://zhuanlan.zhihu.com/p/108152793
Pyradiomics needs to be installed before using this classification model.
-
Use the following command to install pyradiomics or enter their website for other installation methods.
python -m pip install pyradiomics -
We calculate radiomics feature using three different filiters(original, wavelet and LoG). Finally, a total of 1316 features will be obtained.
-
After comparing different radiomics features, we finally use 3D shape-based feature(14 features) to train our classification model because it is the best based on the f1-score of the vghtc 5-fold experiment.
We provide some sample cases from TCGA to familiarize you with this classification model before using your own dataset.
You need to clone our repository and then enter the folder named KidneyCancerClassification.
git clone https://github.com/LinYuXuan-judy/KidneyCancerClassification.git
cd KidneyCancerClassification
You need to run featureExtract.py with the following command to extraction feature. We also provide an environment file (exampleCT.yaml) to set some parameters in pyradiomics, so please don't modify it.
Example:
python3 featureExtract.py samplecase/image samplecase/mask
The result of feature extraction will be stored in a folder named featureResult. We provide two different types of files, one is a csv file and the other is a pkl file, as shown below.
featureResult
├── feature.csv
└── feature.pkl
If you want to use your own dataset, you need to provide two floders, one with all CT images, the other with all mask files of CT images. Then they need to be arranged into a specific structure, as shown below.
- You need to notice that the names of the same case images and masks need to be the same and the data types of images and masks need to be nii.gz files.
samplecase
├── image
│ ├── case_00000.nii.gz
│ ├── case_00001.nii.gz
│ ├── case_00002.nii.gz
│ ├── ...
│
└── mask
├── case_00000.nii.gz
├── case_00001.nii.gz
├── case_00002.nii.gz
├── ...
And running featureExtract.py with the following command.
python3 featureExtract.py path/to/the/image/directory path/to/the/mask/directory
You need to run classification.py with the following command to classify tumors as benign or malignant.
- You don't need to enter the path to the radiomics feature file as we have programmed to fetch its path, so please do not move the location of the radiomics results file.
python3 classification.py
The result of classification will be stored in a folder named classificationResult.
classificationResult
└── classificationResult.csv
We store the results in malignant predict_proba type.
Example:
| case_name | predict_proba |
|---|---|
| case_00000 | 0.561589 |
| case_00001 | 0.231569 |
| case_00002 | 0.895616 |
You need to run classification_lh.py with the following command to classify tumors as low grade or high grade.
python3 classification_lh.py
The result of classification will be stored in a folder named classificationResult.
classificationResult
└── classificationResult_lh.csv
We store the results in high grade predict_proba type.
Example:
| case_name | predict_proba |
|---|---|
| case_00000 | 0.561589 |
| case_00001 | 0.231569 |
| case_00002 | 0.895616 |