PyTorch implementation of toxsmi, a package for toxicity prediction models
from SMILES.
The library itself has few dependencies (see setup.py) with loose requirements.
conda env create -f conda.yml
conda activate toxsmi
pip install -e .In the scripts directory is a training script train_tox.py that makes use
of toxsmi.
Download sample data from the Tox21 database and store it in a folder called data
here.
(toxsmi) $ python3 scripts/train_tox.py data/tox21_train.csv \
data/tox21_score.csv data/tox21.smi data/smiles_language_tox21.pkl \
models params/mca.json test --embedding_path data/smiles_vae_embeddings.pklType python scripts/train_tox.py -h for further help.
Several of our trained models are available via the GT4SD, the Generative Toolkit for Scientific Discovery. See the paper here. We recommend to use GT4SD for inference. Once you install that library, use as follows:
from gt4sd.properties import PropertyPredictorRegistry
tox21 = PropertyPredictorRegistry.get_property_predictor('tox21', {'algorithm_version': 'v0'})
tox21('CCO')The other models are the SIDER model and the ClinTox model from the MoleculeNet benchmark:
from gt4sd.properties import PropertyPredictorRegistry
sider = PropertyPredictorRegistry.get_property_predictor('sider', {'algorithm_version': 'v0'})
clintox = PropertyPredictorRegistry.get_property_predictor('clintox', {'algorithm_version': 'v0'})
print(f"SIDE effect predictions: {sider('CCO')}")
print(f"Clinical toxicitiy predictions: {clintox('CCO')}")ToxSmi uses a self-attention mechanism that can highlight chemical motifs used for the predictions.
In notebooks/toxicity_attention.ipynb we share a tutorial on how to create such plots:
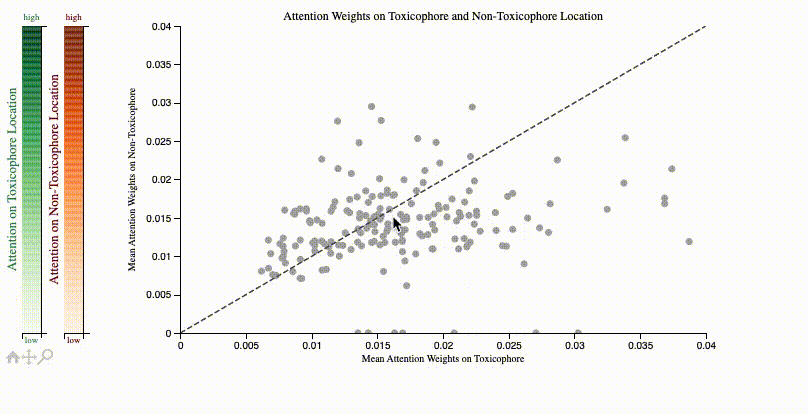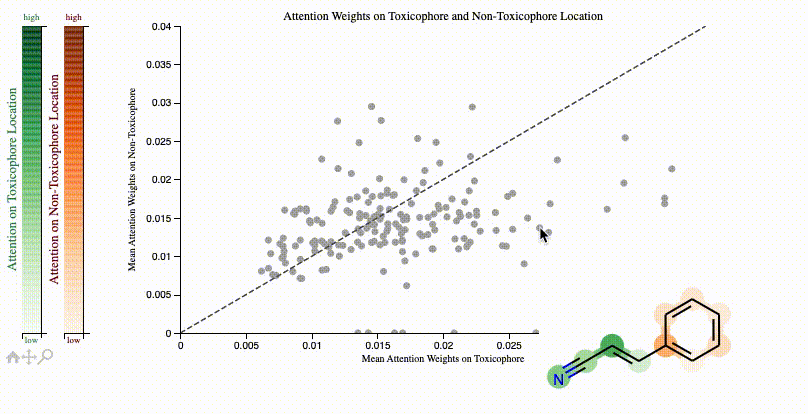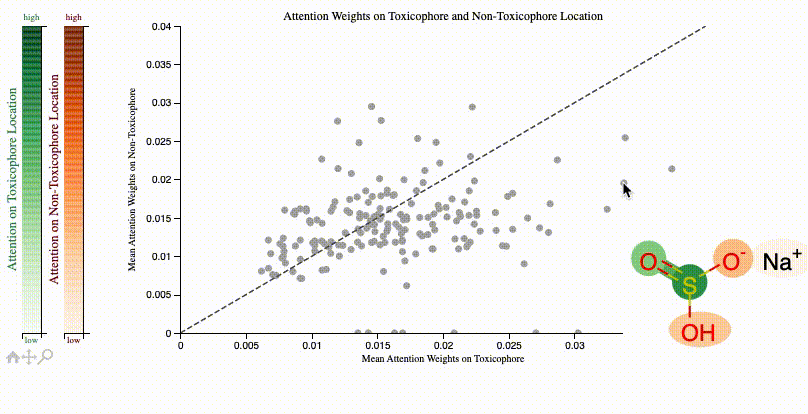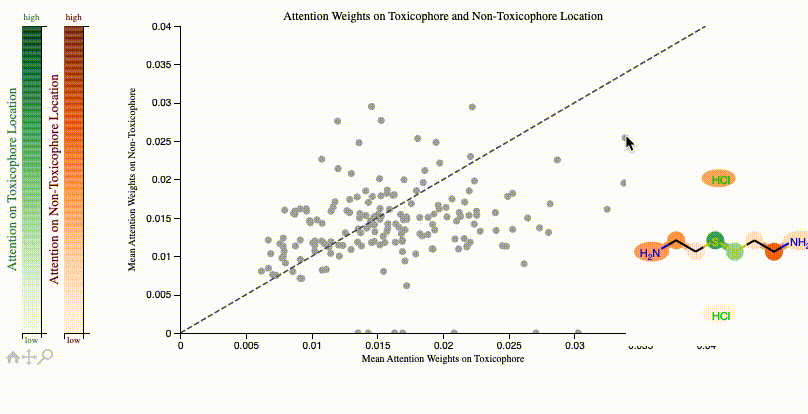
If you use toxsmi in your projects, please (temporarily) cite the following (full paper in review):
@inproceedings{markert2020chemical,
title={Chemical representation learning for toxicity prediction},
author={Markert, Greta and Born, Jannis and Manica, Matteo and Schneider, Gisbert and Rodriguez Martinez, M},
booktitle={PharML Workshop at ECML-PKDD (European Conference on Machine Learning and Principles and Practice of Knowledge Discovery in Databases)},
year={2020}
}