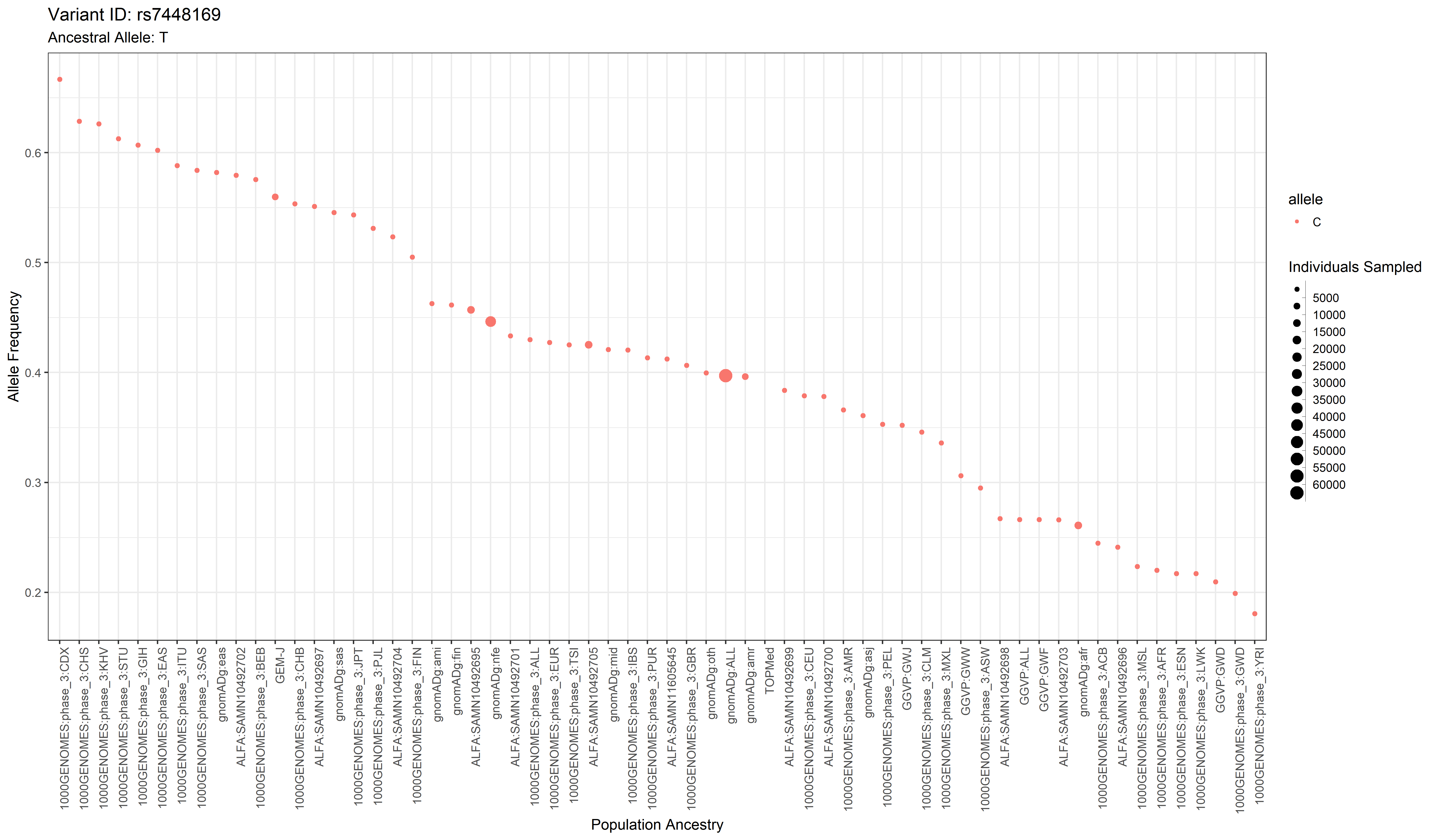
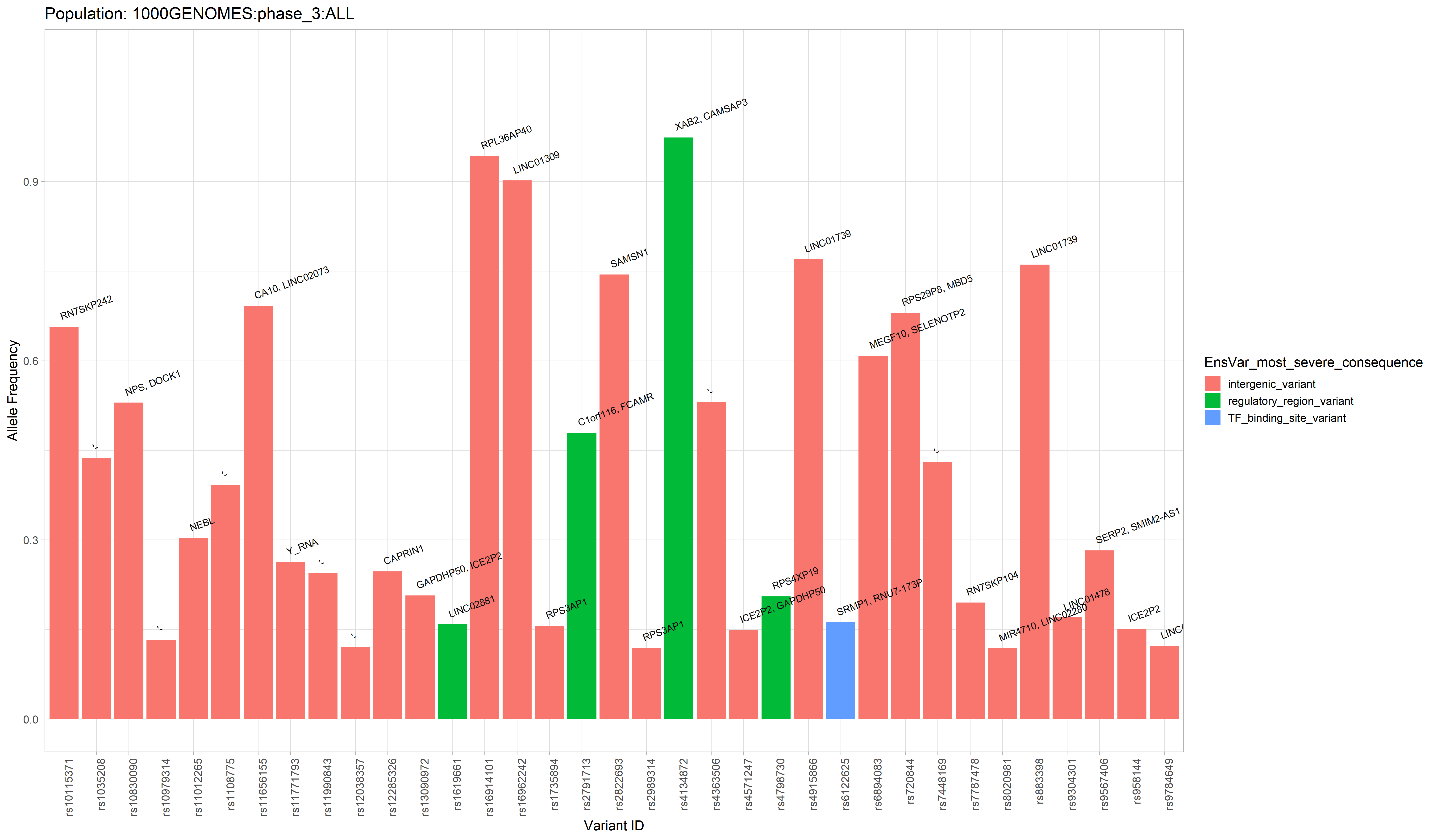
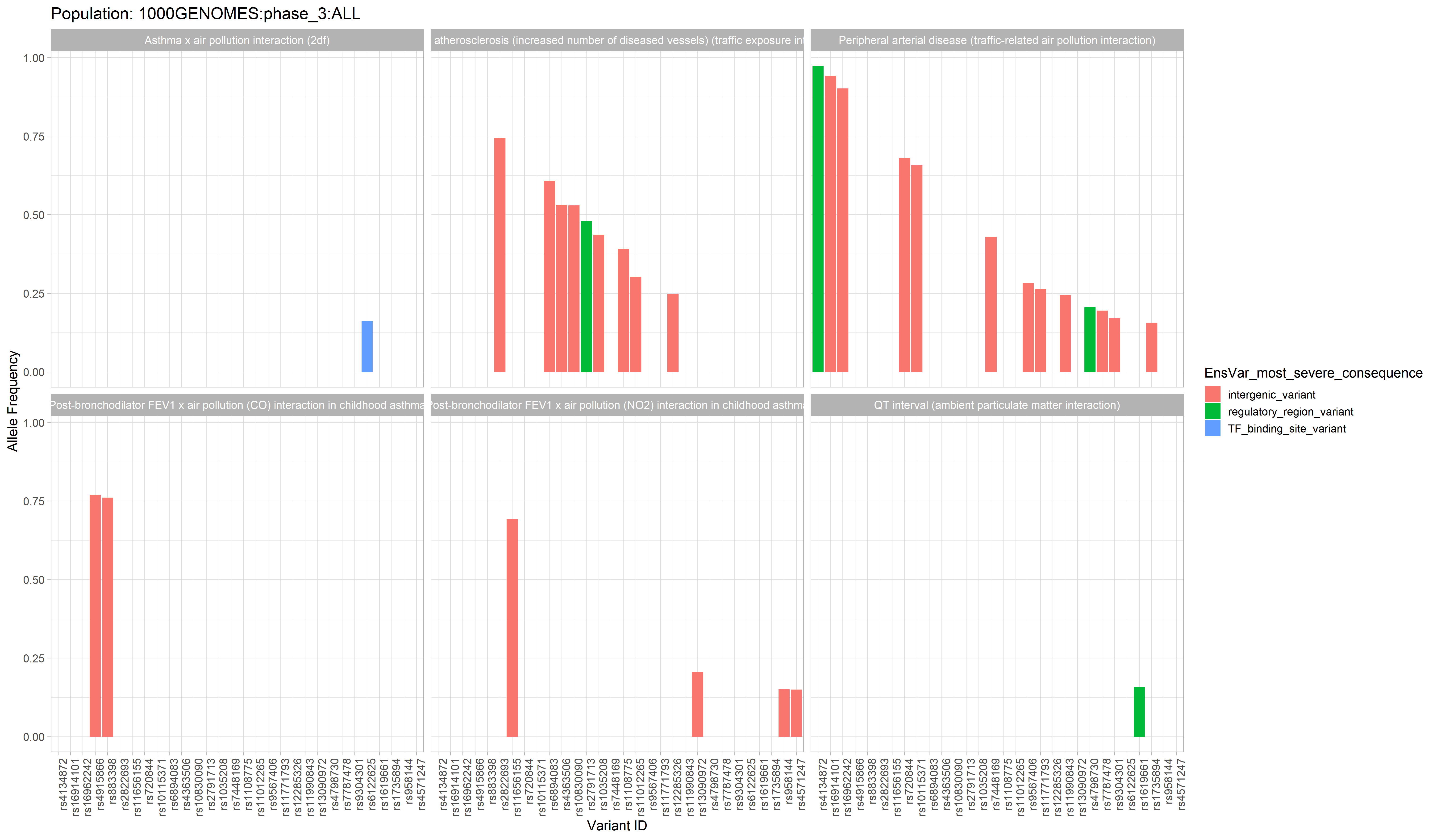
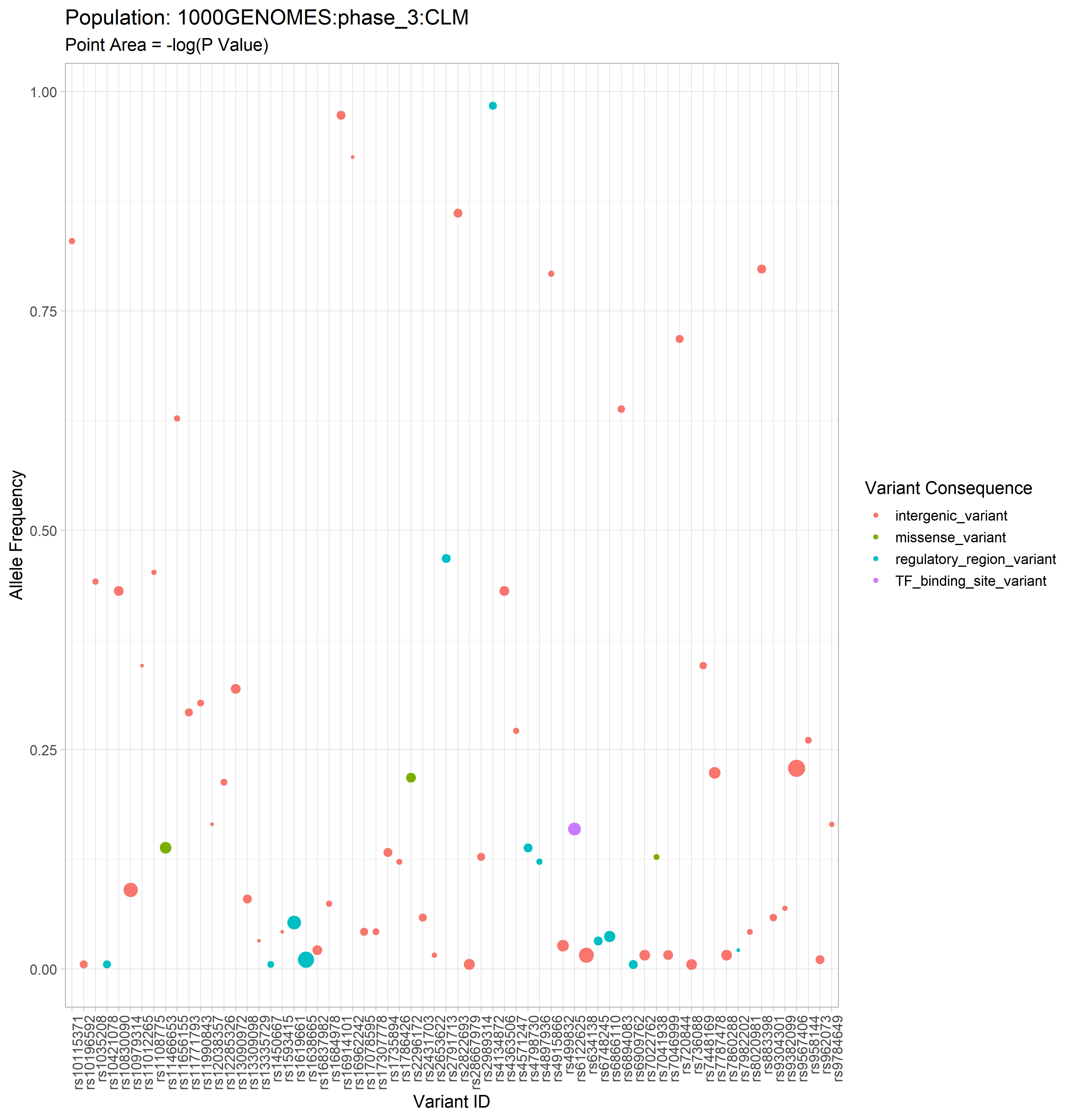
The GWAS catalog (Genome Wide Association Study) is a publically accessible repository of thousands of curated data sets from GWA studies. This package aims to extend its usefulness to researchers interested in connecting phenotypes of interest to genotypes of specific populations.
By integrating population allele frequency data from studies such as The 1000 Genomes Project with GWAS data, phenotypes–traits–of interest which are associated with specific variants–SNPs–can be investigated with respect to populations. The population data is not complete (with respect to all of humanity and all significant sub-populations), but draws from various genomic studies which have been aggregated on Ensembl.
With this R package, a researcher can grab data from the GWAS catalog, and use that data to generate graphs and tables in order to explore the connection between population allele frequencies and alleles associated with a trait of interest.
This package can be installed using the devtools function
install_github()
# Copy this code into your R console in order to download the package.
devtools::install_github("J-T-Nelson/GWASpops.pheno2geno")
There is an independent document which is a full tutorial that explains how this package is meant to be used. The basic workflow needed requires some data retrieval outside of R, thus it is suggested to at least skim the document before attempting to use the package!
Example data available in the “air_pollution” folder which has been used to generate the example plots below.
New users may find this data helpful for learning the various functions in this package, as this data has been persistently used throughout package development and thus is guaranteed to play nicely with all package functions. This very same data was used to create some of the data objects which are included in the package. (More on those data objects in the tutorial)
The data can be downloaded directly from github with the help of a community driven web-app: download-directly.github.io.
Simply go to this website and enter the Github URL of the folder you wish to download: https://github.com/J-T-Nelson/GWASpops.pheno2geno/tree/main/exampleData/air_pollution
If you encounter a bug with the package, have feature requests or wish to discuss this package contact Tanner at jon.tanner.nelson@temple.edu