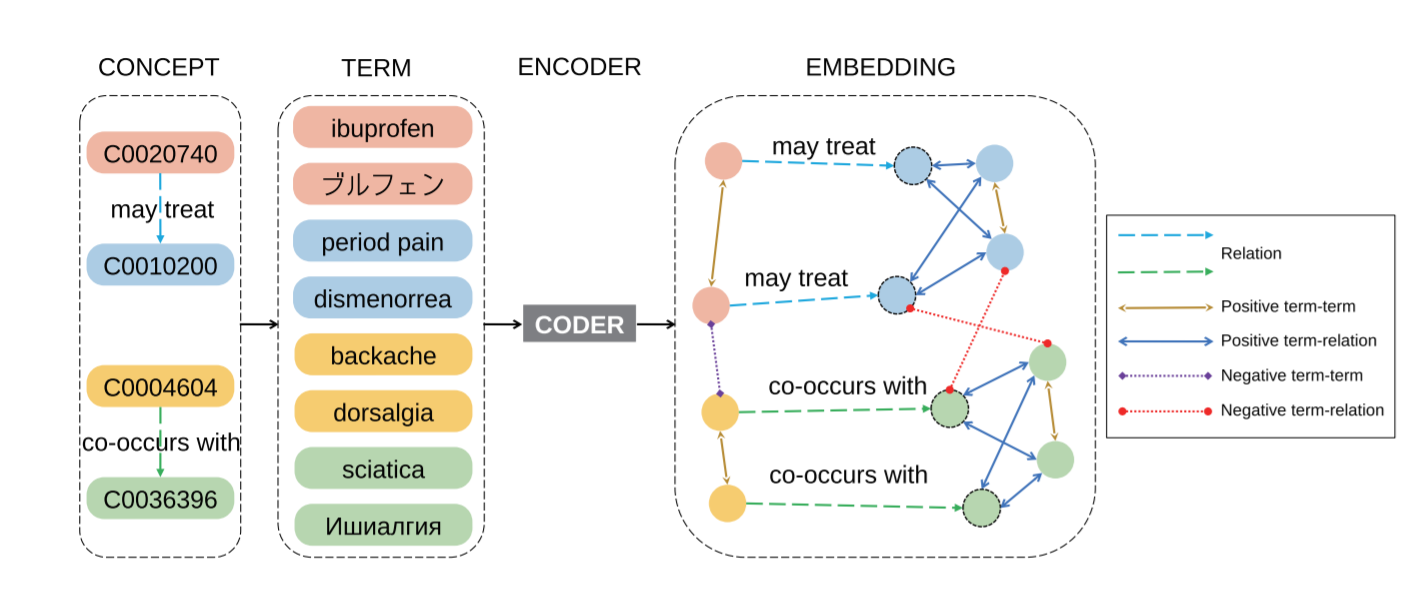
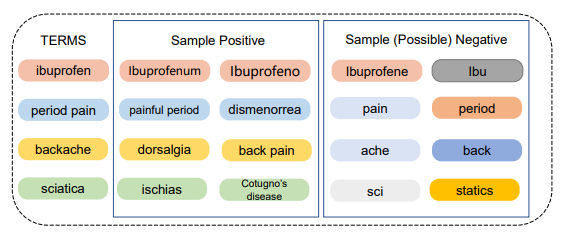
CODER: Knowledge infused cross-lingual medical term embedding for term normalization. Paper
CODER++: Automatic Biomedical Term Clustering by Learning Fine-grained Term Representations. Paper
Models have been uploaded to huggingface/transformers repo.
from transformers import AutoTokenizer, AutoModel
tokenizer = AutoTokenizer.from_pretrained("GanjinZero/UMLSBert_ENG")
model = AutoModel.from_pretrained("GanjinZero/UMLSBert_ENG")English checkpoint: GanjinZero/coder_eng or GanjinZero/UMLSBert_ENG (old name)
English checkpoint CODER++: GanjinZero/coder_eng_pp (with hard negative sampling)
Multilingual checkpoint: GanjinZero/coder_all or GanjinZero/UMLSBert_ALL (discarded old name)
cd pretrain
python train.py --umls_dir your_umls_dir --model_name_or_path monologg/biobert_v1.1_pubmedyour_umls_dir should contain MRCONSO.RRF, MRREL.RRF and MRSTY.RRF. UMLS Download path:UMLS.
from pretrain.load_umls import UMLS
umls = UMLS(your_umls_dir)cd test
python cadec/cadec_eval.py bert_model_name_or_path
python cadec/cadec_eval.py word_embedding_pathDownload the Mantra GSC and unzip the xml files to /test/mantra/dataset, run
cd test/mantra
python test.py
cd test/embeddings_reimplement
python mcsm.pyOnly sampled data is provided.
cd test/diseasedb
python train.py your_embedding embedding_type freeze_or_not gpu_id- embedding_type should be in [bert, word, cui]
- freeze_or_not should be in [T, F], T means freeze the embedding, and F means fine-tune the embedding
@article{YUAN2022103983,
title = {CODER: Knowledge-infused cross-lingual medical term embedding for term normalization},
journal = {Journal of Biomedical Informatics},
pages = {103983},
year = {2022},
issn = {1532-0464},
doi = {https://doi.org/10.1016/j.jbi.2021.103983},
url = {https://www.sciencedirect.com/science/article/pii/S1532046421003129},
author = {Zheng Yuan and Zhengyun Zhao and Haixia Sun and Jiao Li and Fei Wang and Sheng Yu},
keywords = {medical term normalization, cross-lingual, medical term representation, knowledge graph embedding, contrastive learning}
}@misc{https://doi.org/10.48550/arxiv.2204.00391,
doi = {10.48550/ARXIV.2204.00391},
url = {https://arxiv.org/abs/2204.00391},
author = {Zeng, Sihang and Yuan, Zheng and Yu, Sheng},
title = {Automatic Biomedical Term Clustering by Learning Fine-grained Term Representations},
publisher = {arXiv},
year = {2022}
}