This repo contains source code for the scribble-supervised learning paper: Non-iterative Scribble-Supervised Learning for Medical Image Segmentation. October, 2022. [arXiv]
| Dataset | Image Modality | Target Antomy | Scribble | #Images |
|---|---|---|---|---|
| CHAOS [challenge website] | MRI T1-DUAL and T2-SPIR | Liver, Left-kidney, Right-kidney, Spleen | Manual [download link] | 1,917 |
| ACDC [challenge website] | Cine-MRI | Right ventricle, Myocardium, Left ventricle | Manual [download link] | 1,902 |
| LVSC [challenge website] | MRI '2D+time' | Myocardium | Artificial | 29,086 |
Take CHAOS T1 as an example. The following steps transform raw data for training.
- Resample pixel sizes to 1.62$\times$1.62mm$^2$, and then crop or pad the axial view to 256$\times$256pixels.
- Save the 3D volumes slice by slice in .npz format. Each contains an image, label and scribble.
- Split slices into five folds at patient level and store split information in text files.
Specifically, 2D images and five-fold validation text files are stored in ./data/chaos/train_test_split/data_2d and ./data/chaos/train_test_split/five_fold_split respectively.
Dataloaders for loading and augmenting images for network learning is stored in ./datasets/chaos/chaos_dataset.py.
We upload ./data/chaos/train_test_split in Google Drive [here]. Download and unzip the data into your working directory for reproduction.
We train three models for comparison. They are the baseline model, our pacingpseudo model, and the fully-supervised model. The baseline is the UNet backbone penalized by the partial cross entropy loss. Our pacingpseudo uses the siamese architecture to implement the consistency training. Details are described in the paper [arXiv]. The fullly-supervised model is the UNet trained with the cross-entropy loss and Dice loss.
First, create and activate a virtual environment named pacingpseudo with python 3.6.
conda create -n pacingpseudo python=3.6
Then, install necessary packages as follows.
torch=1.7.1
tensorboard=2.7.0
numpy=1.19.5
matplotlib=3.3.4
medpy=0.4.0
simpleitk=2.1.1
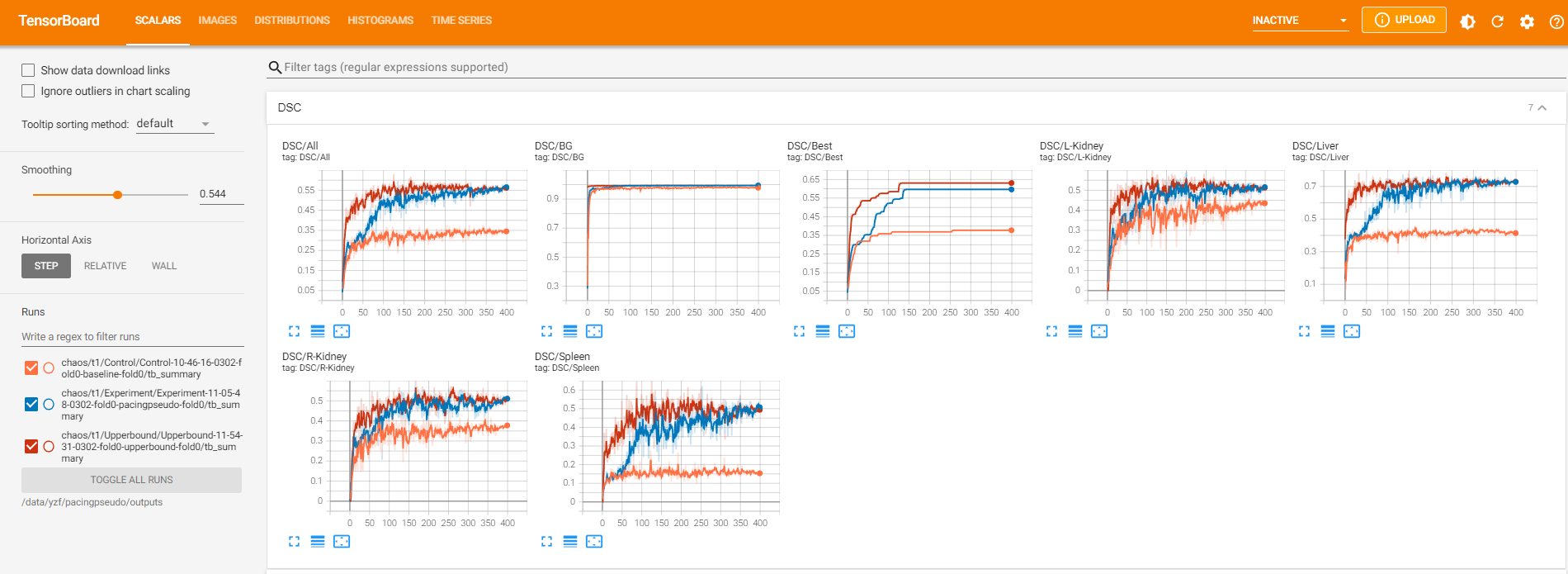
train_chaos.py contains program for training the baseline model and pacingpseudo model. upperbound_chaos.py is for training the fully-supervised model. At the end of each training epoch, we compute Dice similarity coefficients on the validation dataset and plot them using TensorBoard.
To run the training programs, we have to input configuration parameters. Below is the command line for learning the baseline model on CHAOS T1.
python train_chaos.py --gpu=0 --session=Control --tag=baseline-fold0 --fold=0 --modality=t1
The pacingpseduo model needs additional configurations. Its training command line is as follows.
python train_chaos.py --gpu=0 --session=Experiment --tag=pacingpseudo-fold0 --fold=0 --modality=t1 --do_loss_ent --do_decoder_consistency --do_aux_path --do_memory
The upperbound model uses upperbound_chaos.py to train. Below is the command line to run upperbound_chaos.py.
python upperbound_chaos.py --gpu=0 --session=Upperbound --tag=upperbound-fold0 --fold=0 --modality=t1
Training outputs are stored in ./outputs. They include logging files, model checkpoints, and tensorboard files.
Tensorboard files monitor loss values, validation results, intermediate predictions during training. The following command calls the tensorboard interface.
tensorboard --logdir=./outputs --port=6007 --bind_all
Then, on your browser, type localhost:6007 to watch the interface.
Inference.py is designed to compute evaluation metrics given a model parameter checkpoint. We use the final checkpoint in our experiments. The metrics includes the Dice similarity coefficients (DSC) and the 95-th percentile of Hausdorff distance (HD95). Roughly speaking, DSC quantifies relative overlap and HD95 quantifies boundary distance. Here is a lecture on Hausdorff distance between convex polygons.
The following command uses the pacingpseduo model to inference validation metrics. We load model parameters at the final checkpoint by default.
python inference.py --gpu=0 --dataset=chaost1 --fold=0 --checkpoint_file=./outputs/chaos/t1/Experiment/Experiment-11-05-48-0302-fold0-pacingpseudo-fold0
Inference outputs are stored in ./outputs/inference.
We summarize results in DSC and HD95. HD95 is measured in millimeters (mm). Higher is better for DSC and lower is better for HD95.
Below are the results of five-fold validation. Specifically, in each fold, we first compute the average score of each anatomy over patients, which results in
Displayed below are experimental results on CHAOS T1.
DSC results on CHAOS T1| Models | Fold 0 | Fold 1 | Fold 2 | Fold 3 | Fold 4 | Overall Average |
|---|---|---|---|---|---|---|
| Baseline | 0.3452 | 0.3732 | 0.4149 | 0.4578 | 0.4234 | 0.4029 |
| PacingPseudo (ours) | 0.5633 | 0.6583 | 0.6751 | 0.7030 | 0.7283 | 0.6656 |
| Fully-supervised | 0.5632 | 0.6162 | 0.6774 | 0.7662 | 0.7743 | 0.6795 |
| Models | Fold 0 | Fold 1 | Fold 2 | Fold 3 | Fold 4 | Overall Average |
|---|---|---|---|---|---|---|
| Baseline | 60.79 | 74.24 | 41.51 | 34.67 | 48.36 | 51.91 |
| PacingPseudo (ours) | 17.20 | 17.96 | 12.11 | 10.94 | 10.44 | 13.73 |
| Fully-supervised | 16.29 | 25.04 | 12.86 | 9.55 | 9.09 | 14.57 |
The output data ./outputs , including traning and inference outputs, are uploaded in Google Drive [here].
@article{yang2024non,
title={Non-iterative scribble-supervised learning with pacing pseudo-masks for medical image segmentation},
author={Yang, Zefan and Lin, Di and Ni, Dong and Wang, Yi},
journal={Expert Systems with Applications},
volume={238},
pages={122024},
year={2024},
publisher={Elsevier}
}