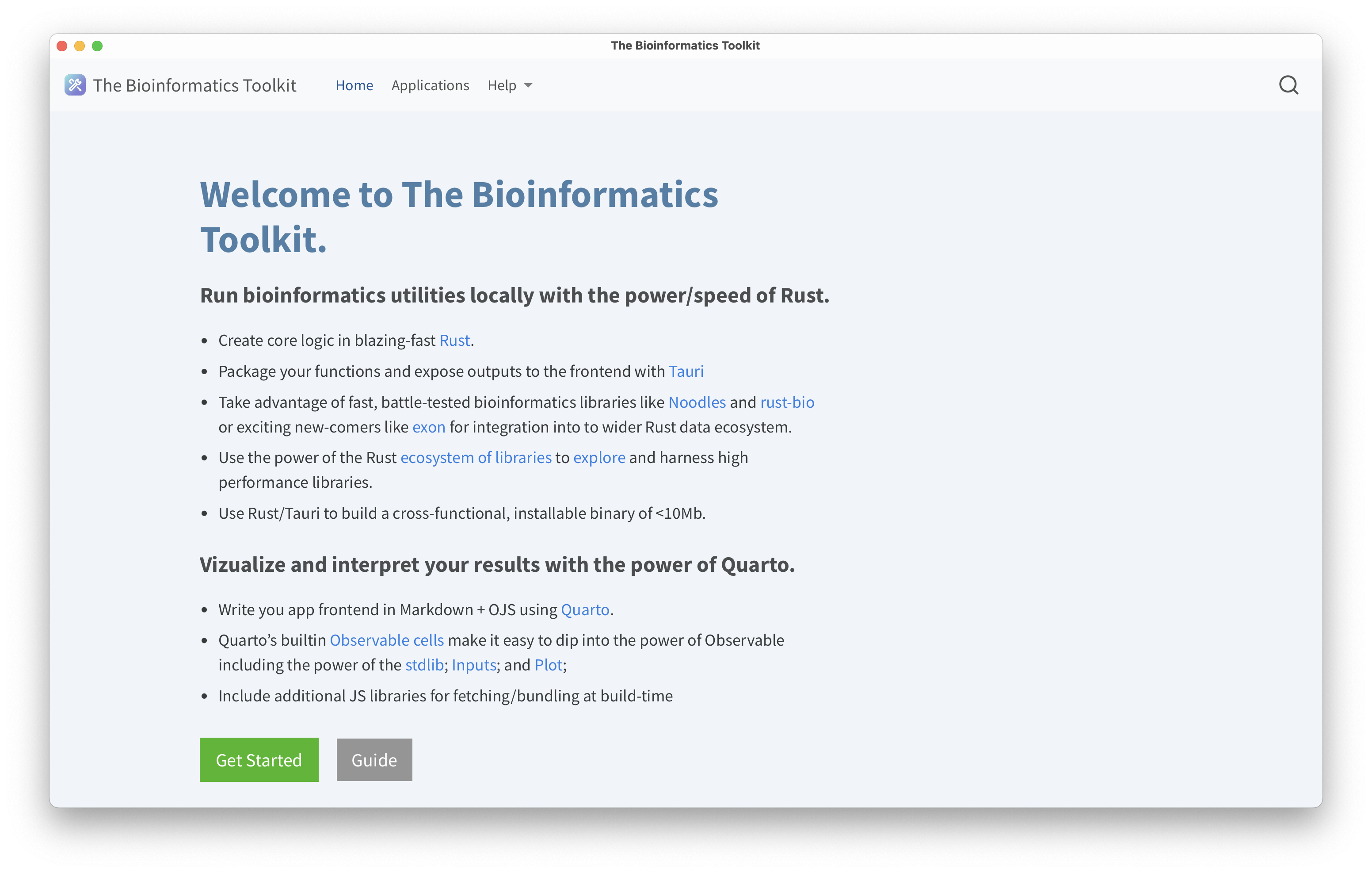
RUST-backed utilities for bioinformatic data processing.
The fastest way to get started it to download the applications found in the Releasehttps://github.com/zachcp/bioinformaticstoolkit/releases section. This project aims to demonstrate how the Rust toolchain enables efficient cross-platform support for high-performance applications. By using Tauri you can write the entire frontend in any tool that compiles to HMLT+Javascript, in this case I used Quarto to take advantage of its simple composition (its mostly markdown +yaml) as well as it's built-in use of the observable runtime.
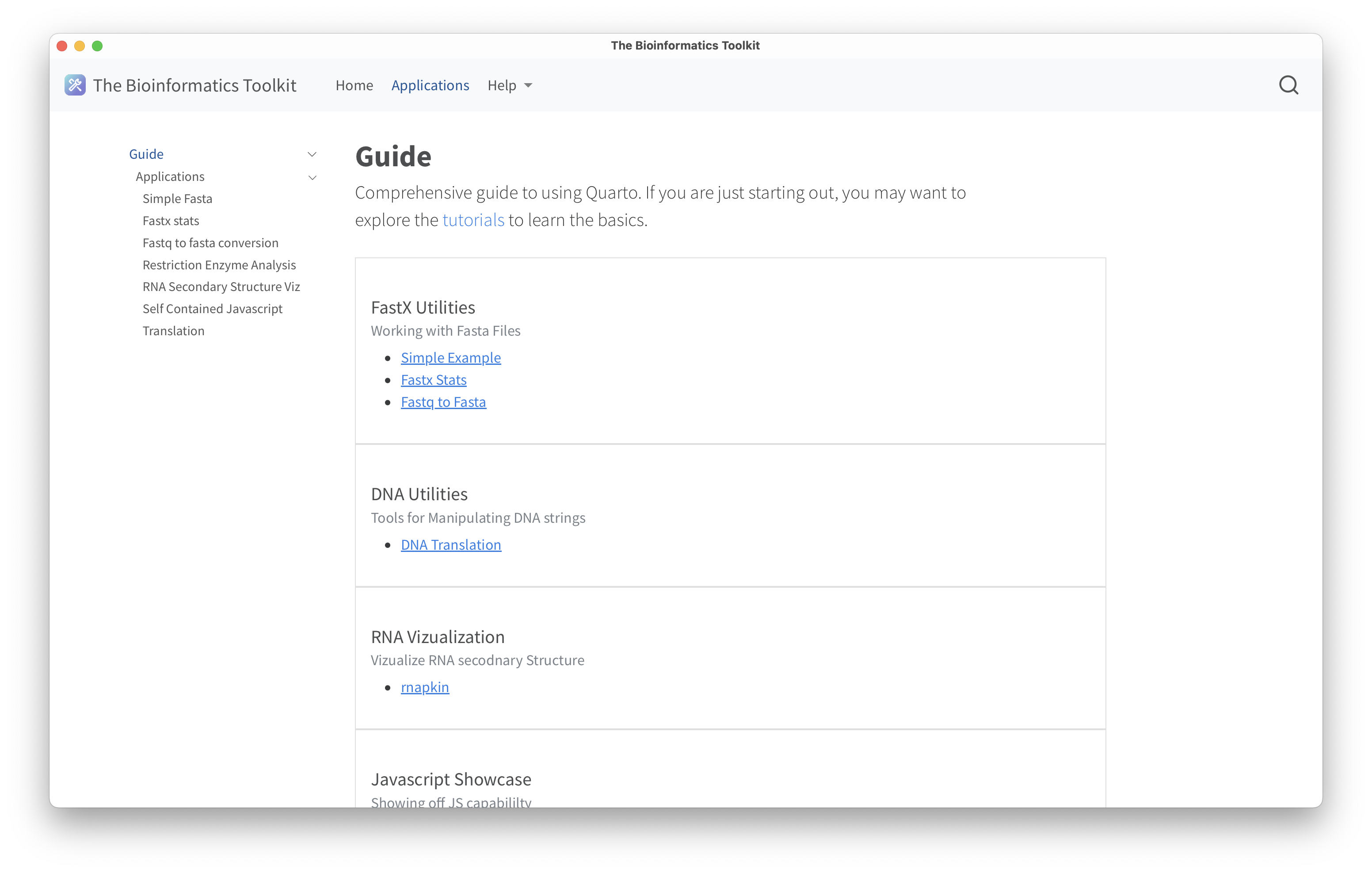
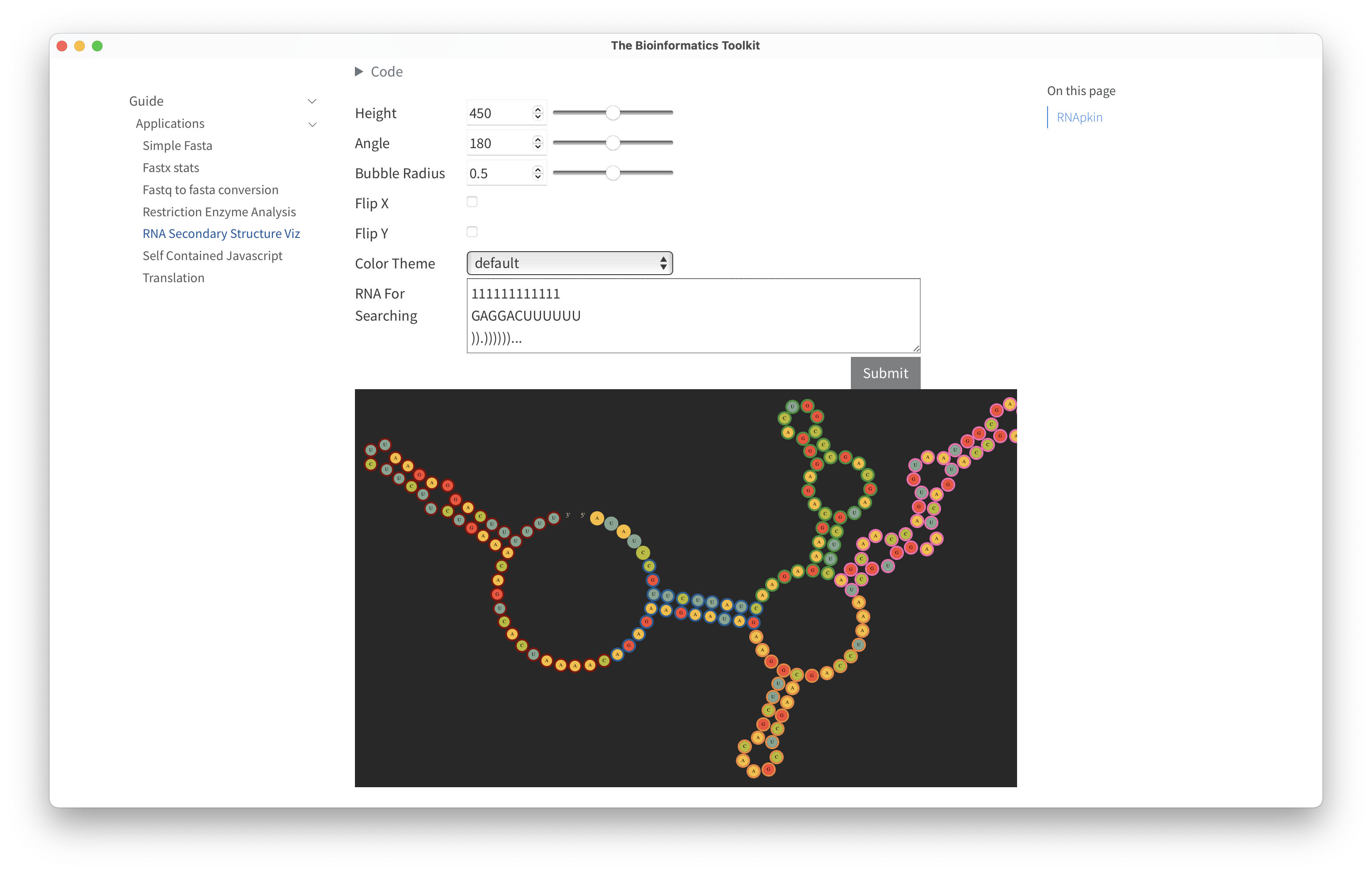
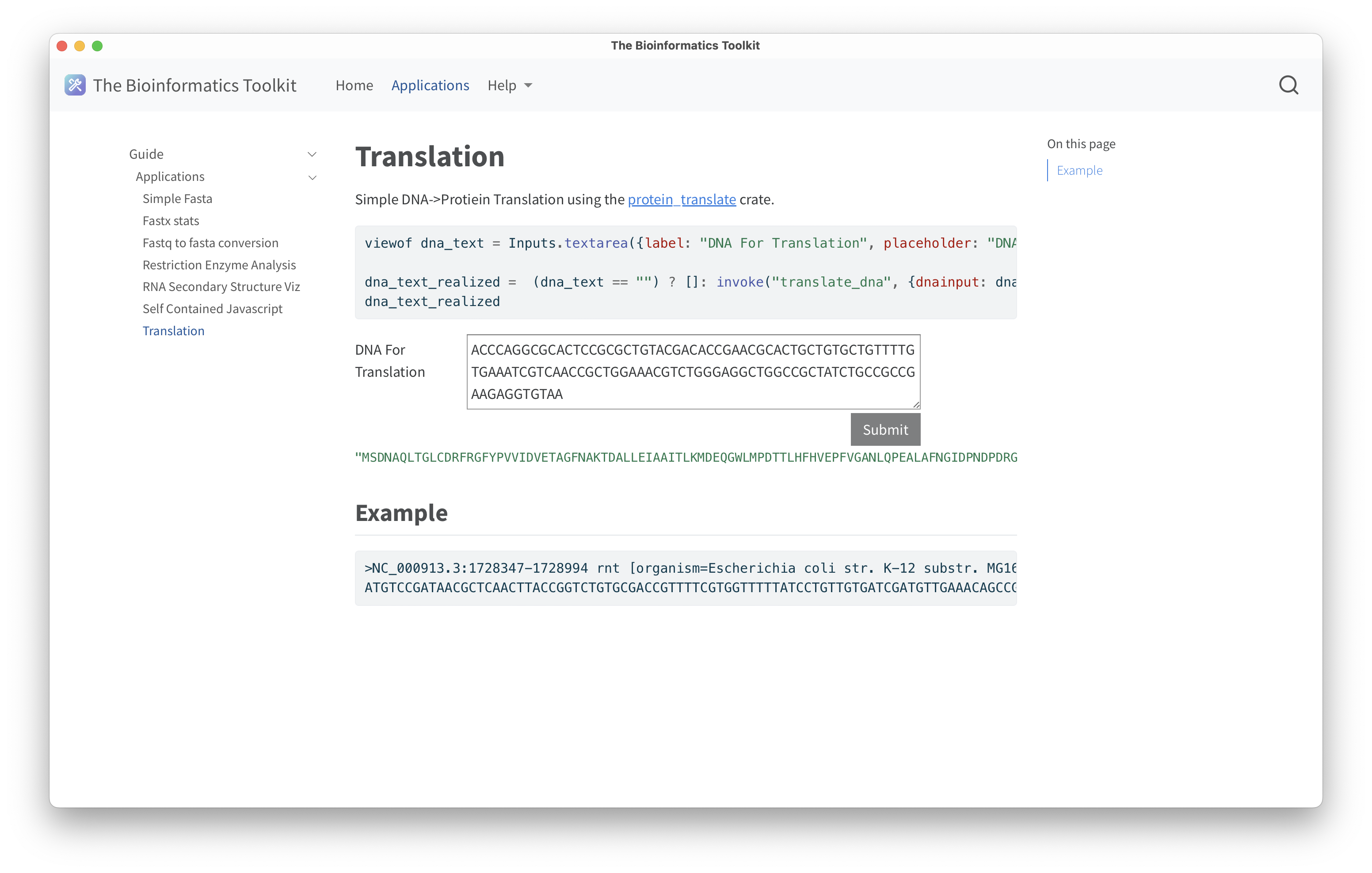
Below are screenshots of a native application demonstrating the home page, the guide page, an example RNA secondary strucutre visualization using rnapkin;statistics of a fasta file including a histrogram of sequence lengths using noodles for IO; and DNA translation using the protein_translation crate.
# assuming quarto and cargo are installed and on your path.
git clone https://github.com/zachcp/bioinformaticstoolkit.git
cd bioinformaticstoolkit
# install the tauri cli
cargo install tauri-cli
# add cargo bind dir to the path
export PATH=$PATH:~/.cargo/bin/
# to develop
cargo-tauri dev
# to package. this build is ~8MB.
cargo-tauri build
# to test
cd src-tauri && cargo test
# or verbose
cd src-tauri && cargo test -- --nocapture
FASTX:
- convert fasta to fastq
- basic stats of fasta/fastq
- histrogram of read lengths (possibly set max number)
- merge PE reads // split interleaved
- splitting into multiple files ( create directory ?)
- filter-fastx length // quality
- sample the fasta/x files
- plot: length x quality metrics ( optional hexagon plots )
- plot: coverage by location.
GFA:
- Utilites from GFATK including filtering
- GFAStats
DNA Analysis:
- Digestability of DNA sequences:
- Search for RE locations
- Other Patterns to Avoid
- Data: Standard RE enzymes
- Plot: Genome View of RE sites.
- Global view of Palettes and coding types
- Insilico PCR: https://github.com/dlesl/pcr
- Clonifier: https://github.com/dlesl/clonifier
- Phenogram
- Pangenome TK: https://github.com/GeneDx/pgr-tk (cdep in the build)
- RE digest and assembly calculations
VCF:
- convert
- concat
- split
RNA Secondary Structure:
- RNApkin https://lib.rs/crates/rnapkin
rna-seq: - [ ] gencounts https://github.com/NKI-GCF/gensum - [ ] rust-lapper https://crates.io/crates/rust-lapper
Taxonomy:
- load and display a tree file
- load and display kraken
- load and display bracken
Peptides and Proteomics:
Rust Software:
Miscelleaneous:
- Genome Card: e.g viz with global genome statistics.
- Genome name, overview, produces compounds
- Utilities for Codons
- VCF plotein
- ASGArt (cdep in the build)
- UDON
- GFAESTUS (c++ dep )
- BioSeq
- 10x Genomics Rust
- fq parser
- fastats
- fqmerge
- ggcat
- light motif
- liftover with crusmapr
- exon
- phylogeny # not much action
- chemical Reaction networks
- gb-io
- charming - a nive gui library
- met map
- barcode counter
- hpo
- nanopore read assessment: https://lib.rs/crates/nanoq#readme-read-report
- niffler
- OBO Validatio
- rustyms
- preotienogenic
- rdkit
- bigwig2bam
- Plasmapr: https://github.com/BradyAJohnston/plasmapR
- flate2use flate2::read::MultiGzDecoder;
- bio_streams
- Streaming iterators for bioinformatics data