0. What is this?
This is a solution to the EBM-NLP task proposed in this ACL 2018 publication by Benjamin Nye et al.
The method is Named Entity Recognition (NER) with BioELMo + CRF under PyTorch implementation.
1. Preparation
- Clone this repository:
$ git clone https://github.com/iBotamon/ebmnlp.git- Activate Virtual Environment:
$ cd ebmnlp
$ python -m venv .
$ source bin/activate- Install necessary packages:
$ pip install --upgrade pip
$ pip install -r requirements.txt- Download the following files:
- Save Model checkpoint in
models/ebmnlp_bioelmo_crf. - Save BioELMo weights in
models/bioelmo. - Save BioELMo options in
models/bioelmo. - Save BioELMo vocabulary in
models/bioelmo.
Instead, you can also download them by running this:
$ bash get_pretrained_models.sh2. How to use BioELMo + CRF model
2-1. Use via command line
-
Prepare text file that contains an RCT abstract (e.g., sample.txt).
-
Run like this:
$ python ebmnlp.py TEXT_FILE_NAME- NER tagging result will be returned in a standard output:
I-I Remdesivir
O in
I-P adults
I-P with
I-P severe
I-P COVID-19
I-P :
O a
O randomised
O ,
O double-blind
O ,
O placebo-controlled
O ,
O multicentre
- If you wish to get the result as a file, run like this:
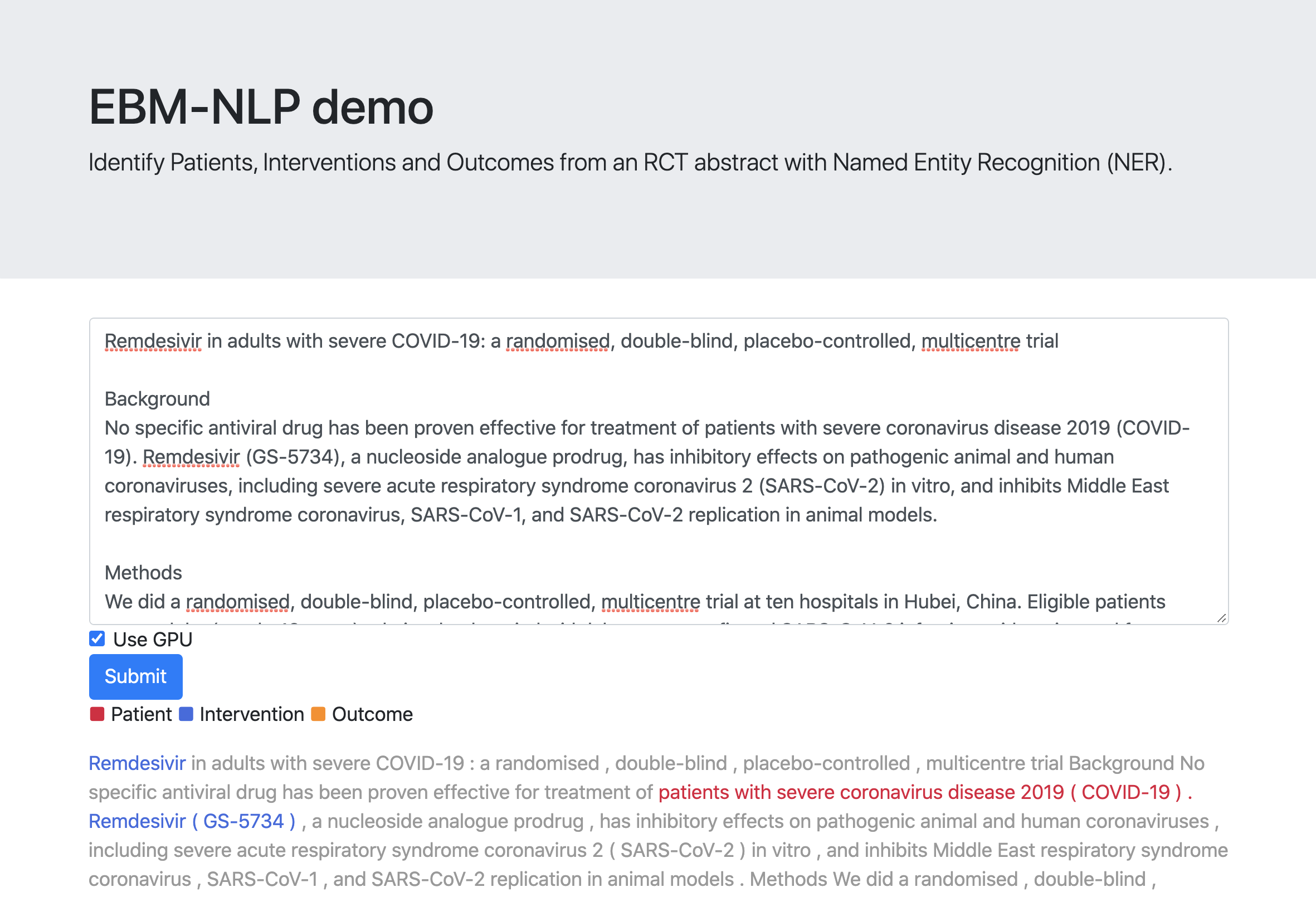
$ python ebmnlp.py TEXT_FILE_NAME OUTPUT_FILE_NAME2-2. Use via Web browser
- Run this:
$ bash run_flask.sh
-
Access to
localhost:5000via your Web browser. -
You can use the PIO identification system interactively.
3. How to train BioELMo + CRF model yourself
-
Prepare EBM-NLP dataset
ebm_nlp_1_00.tar.gzfrom the repository by the authors. -
Extract
ebm_nlp_1_00.tar.gzin theofficialdirectory like this:
- models
- templates
- official
└ ebm_nlp_1_00
└ annotations
└ ..
└ documents
└ ..
- Run this:
$ python ebmnlp_bioelmo_crf.pyYou can specify CUDA device number like this:
$ python ebmnlp_bioelmo_crf.py --cuda 3