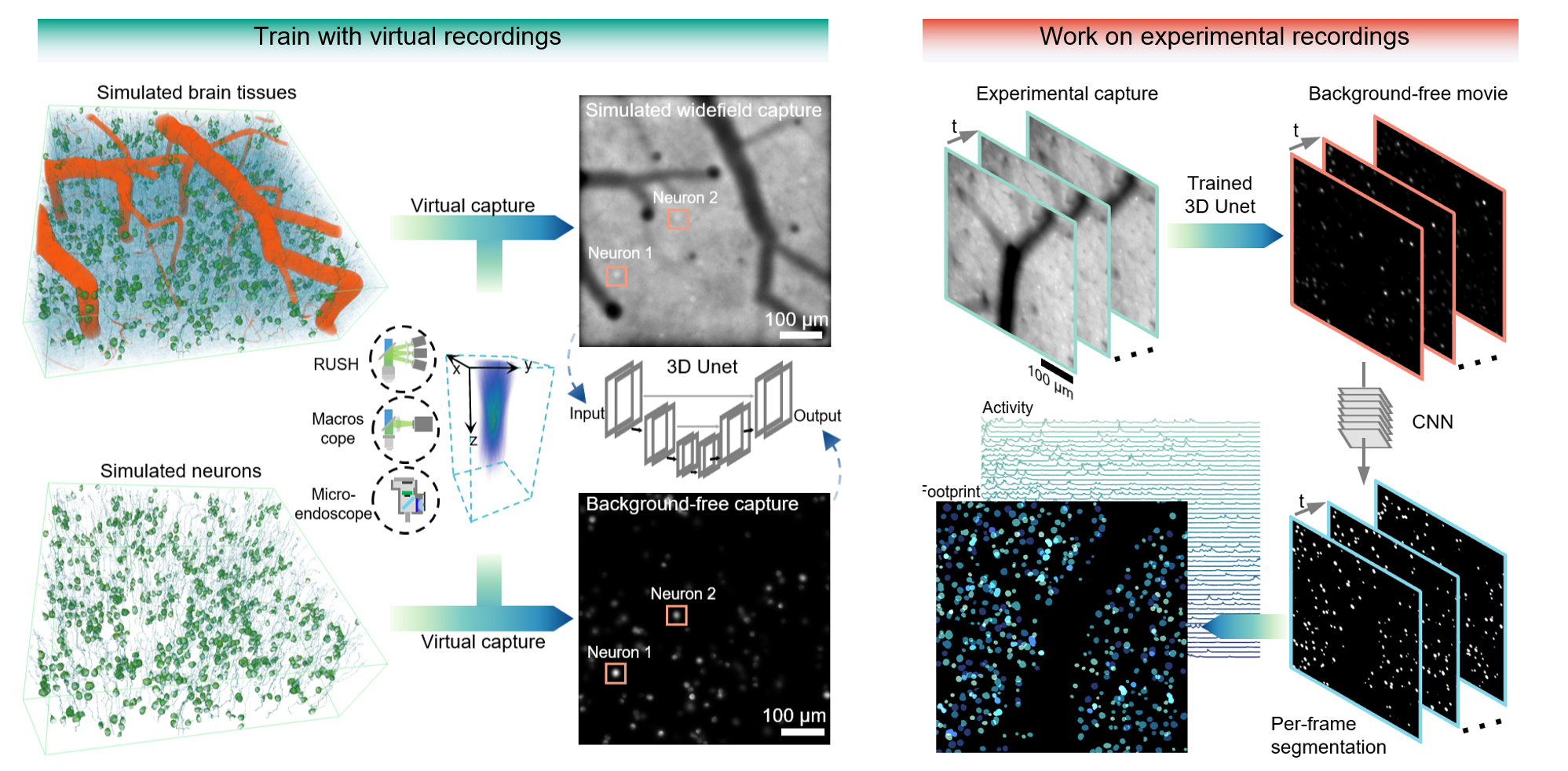
Widefield imaging provides optical access to multi-millimeter fields of view in video rate, but cell-resolved calcium imaging has mostly been contaminated by tissue scattering and calcium inference suffers multiple times slower processing speed than two-photon microscope in the same scale. We present a deep widefield neuron finder (DeepWonder), which is fueled by numerical recordings but effectively works on experimental data with an order of magnitude of speed improvement and improved precision and accuracy. With DeepWonder, widefield calcium recordings can be quickly converted into background-free movies, and neuronal segments and activities then be demixed out. For more details and results, please see our companion paper titled "Rapid widefield neuron finder driven by virtual calcium imaging data".
This tutorial will show how DeepWonder removes backgrounds in widefield captures and further segments neurons.
If you encounter issues following the tutorial below, you can try the pipeline in the 'alternative' folder. There's a separate README file within that folder to guide you.
- Ubuntu 16.04
- Python 3.6
- Pytorch >= 1.3.1
- NVIDIA GPU (24 GB Memory) + CUDA
- Create a virtual environment and install some dependencies.
$ conda create -n deepwonder_env python=3.6
$ source activate deepwonder_env
$ pip install -q torch==1.10.0
$ pip install -q torchvision==0.8.2
$ pip install DWonder
$ pip install -q opencv-python==4.1.2.30
$ pip install -q tifffile
$ pip install -q scikit-image==0.17.2
$ pip install -q scikit-learn==0.24.1
$ git clone git://github.com/yuanlong-o/Deep_widefield_cal_inferece
$ cd DeepCAD/DeepWonder/
We upload three demo data on Google drive: synthetic widefield data by NAOMi1p code , cropped RUSH data and widefield data jointly with two-photon ground truth. To run the demo script, those data need to be downloaded and put into the DeepWonder/datasets folder. We upload a trained background removing model and a train neuron segmentation model. And you can get the background removing results of these three demo data on Google drive: synthetic widefield data by NAOMi1p code , cropped RUSH data and widefield data jointly with two-photon ground truth. Especially, We upload the two-photon reference data for widefield demo data.
(5/30/2022) We further upload over 50Gb paired hybrid data on Google drive (https://drive.google.com/drive/folders/1OBcQUY-vsIPljSBChFfn-zqAYtYvDZ4A?usp=sharing),including:
• Data that are captured across brain wide (widefield + NA 0.27 2p).
• Data that are distributed in different cortical depths (widefield + NA 0.27 2p).
• High-NA validation data that are captured in multiple cortical regions (widefield + NA 0.60 2p).
Run the script.py to analyze the demo data.
$ python script.py
The output from the demo script can be found in the DeepWonder/results folder. For example, the background removed movie is in the DeepWonder/results/XXXX/RMBG folder, the neuron footprint masks are in the DeepWonder/results/XXXX/f_con folder and stored in a .tif file, and the activities are in the DeepWonder/results/XXXX/mat folder as a .mat file.
Since the uploaded pretrained DeepWonder was trained on the synthetic data from NAOMi1p (see following instructions) under RUSH modalities (the specific parameters are in NAOMi1p\config\RUSH_ai148d_config.m) which might be different from your acquisition specifications, it is highly recommended to follow the re-train instructions down below to get the best performances. As an alternative option, new data can be resized into 0.8 um pixel size to mimic the modality in the pretrained situation and put in the DeepWonder/datasets folder. After that, DeepWonder is ready to go as suggested previously.
We have stored our data in Google Colab, which is a free Jupyter notebook environment that requires no setup and runs entirely in the cloud. A demo notebook with full processing of DeepWonder on several demo datasets (including NAOMi1p virtual datasets, cropped RUSH datasets, and two-photon validation datasets, all mounted to the Google Colab using Google Drive) is available through Colab via
.
By copying that notebook to your online server (like Colab), you can also process linked data in the online fashion.
DeepWonder relies on a highly realistic simulation of widefield capture for training a network that removes widefield background. We referred to the Neural Anatomy and Optical Microscopy (NAOMi) package (https://www.sciencedirect.com/science/article/pii/S0165027021001084) to populate the brain tissue with multiple blood vessels, somata, axons, and dendrites. We further modified the NAOMi pipeline such that it could faithfully simulate data acquisition of one-photon excitations with one-photon excitation model and related noise model, which we termed as NAOMi1p. The virtual widefield data by NAOMi1p can be affected by optical parameters, illumination powers, and other parameters like protein concentration. The full key parameters are listed as
- Optical parameters of the microscope: NA, FOV, FN, immersion medium
- Indicator parameters: expression level, indicator types (e.g. GCaMP6 or GCaMP7)
- Imaging parameters: session length, frame rate, illumination powers, and imaging depth
- (optional) Vessel dilation parameters: dilation dynamics, amplitude, and anisotropy across the FOV.
All those parameters should be adjusted based on a specific system. As an example, run NAOMi_1p_single.m to generate a 750 x 750 x 1000 frame virtual widefield recording (mov_w_bg.tiff) along with a background free pair (mov_wo_bg.tiff). To generate multiple training pairs, run NAOMi_1p_loop.m which nests a for loop to generate data.
Put the backaground removed movie generated by NAOMI1p to the DeepWonder/datasets/XXXX/GT folder, and the paired background contaminated movie to the DeepWonder/datasets/XXXX/Input folder. After preparing the data, run the script_train_RMBG.py to train the background removal network.
$ source activate deepwonder_env
$ python script_train_RMBG.py train
The trained removing background model will show up in DeepWonder/RMBG_pth folder. We also provide pre-trained models to speed up the fine tuning process. This model is fully trained on a large number of simulated data with different characteristics and can be used as the initialization of the background removal network model. Run the script_train_RMBG.py to train the background removal network with pretrain model initilaization.
$ python script_train_RMBG_conti.py train
Put the backagrond removed movie generated to the DeepWonder/datasets/XXXX/image folder as inputs, and the corresponding segmented masks to the DeepWonder/datasets/XXXX/mask folder as labels. A good option to get the mask data is to binarize simulated neurons for each of frames (individual neurons are in the vol_out structure from NAOMi1p output). Alternatively, running other two-photon segmentation tools (like CaImAn https://github.com/flatironinstitute/CaImAn) to get segments can also work but probably with performance dropping. Run the script_train_SEG.py to train the neuron segmentation network.
$ source activate deepwonder_env
$ python script_train_SEG.py train
The trained neuron segmentation model will show up in DeepWonder/SEG_pth folder.
Zhang, Y., Zhang, G., Han, X. et al. Rapid detection of neurons in widefield calcium imaging datasets after training with synthetic data. Nat Methods (2023). https://doi.org/10.1038/s41592-023-01838-7
We are pleased to address any questions regarding the above tools through emails (zhanggx19@mails.tsinghua.edu.cn or yuanlongzhang94@gmail.com).