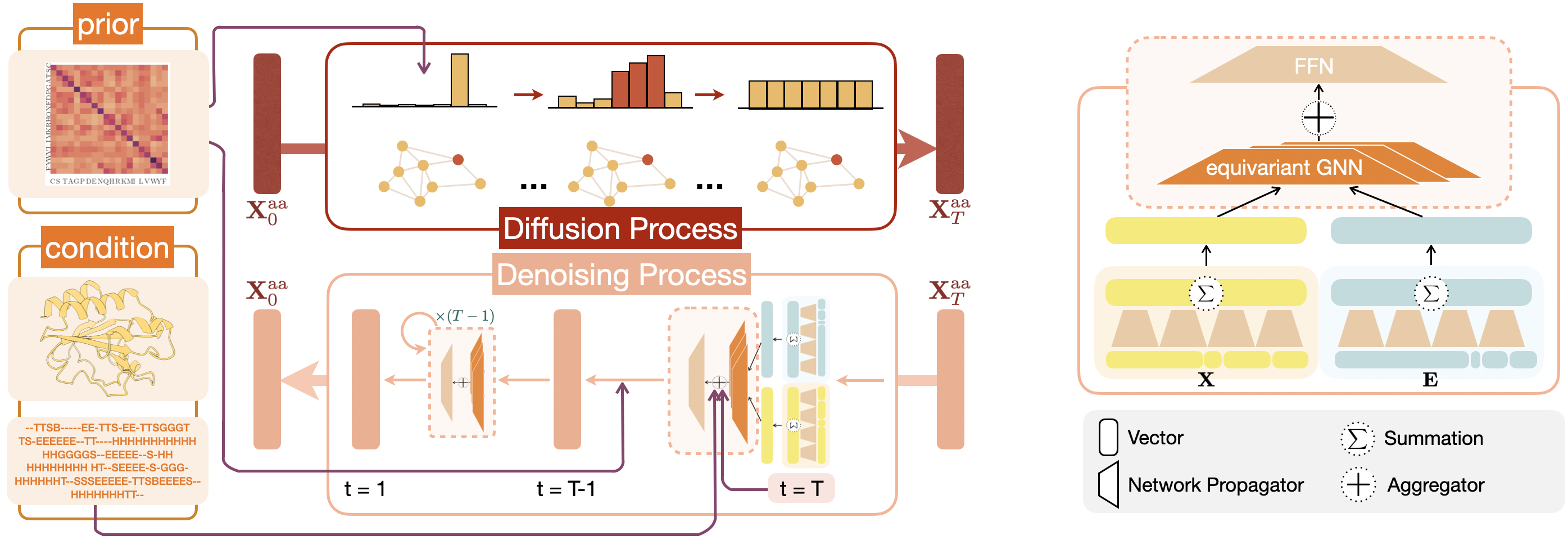
Implementation for "Graph Denoising Diffusion for Inverse Protein Folding" arxiv link.
To install requirements:
conda env create -f environment.yml
Like denoising-diffusion-pytorch, there is a brief introduction to show how this discrete diffusion work.
import sys
sys.path.append('diffusion')
import torch
from torch_geometric.data import Batch
from diffusion.gradeif import GraDe_IF,EGNN_NET
from dataset_src.generate_graph import prepare_graph
gnn = EGNN_NET(input_feat_dim=input_graph.x.shape[1]+input_graph.extra_x.shape[1],hidden_channels=10,edge_attr_dim=input_graph.edge_attr.shape[1])
diffusion_model = GraDe_IF(gnn)
graph = torch.load('dataset/process/test/3fkf.A.pt')
input_graph = Batch.from_data_list([prepare_graph(graph)])
loss = diffusion_model(input_graph)
loss.backward()
_,sample_seq = diffusion_model.ddim_sample(input_graph) #using structure information generate sequenceMore details can be found in the jupyter notebook
Here is an ablation study of two key parameters, step and diverse, in the ddim_sample function used to get improved results presented in the paper. The following results were computed after 50 ensemble runs. One can find how to do ensembles in the jupyter notebook.
| Step | Recovery Rate | Perplexity | Single Sample Recovery Rate |
|---|---|---|---|
| 500 | 0.5341 | 4.02 | 0.505 |
| 250 | 0.5370 | 4.06 | 0.4679 |
| 100 | 0.5356 | 4.98 | 0.4213 |
| 50 | 0.4827 | 8.02 | 0.3745 |
| Step | Recovery Rate | Perplexity | Single Sample Recovery Rate |
|---|---|---|---|
| 500 | 0.5342 | 4.02 | 0.505 |
| 250 | 0.5373 | 4.12 | 0.4741 |
| 100 | 0.5351 | 7.43 | 0.5016 |
| 50 | 0.4999 | 16.74 | 0.4736 |
| Step | Recovery Rate | Perplexity | Single Sample Recovery Rate |
|---|---|---|---|
| 500 | 0.5286 | 4.08 | 0.5022 |
| 250 | 0.5292 | 4.13 | 0.4325 |
| 100 | 0.5329 | 5.28 | 0.4222 |
| 50 | 0.5341 | 5.91 | 0.4212 |
| Step | Recovery Rate | Perplexity | Single Sample Recovery Rate |
|---|---|---|---|
| 500 | 0.5286 | 4.08 | 0.5022 |
| 250 | 0.5273 | 4.09 | 0.4357 |
| 100 | 0.5238 | 9.49 | 0.5095 |
| 50 | 0.5285 | 15.53 | 0.5113 |
- Our codebase for the EGNN models and discrete diffusion builds on EGNN, DiGress. Thanks for open-sourcing!
If you consider our codes and datasets useful, please cite:
@inproceedings{
yi2023graph,
title={Graph Denoising Diffusion for Inverse Protein Folding},
author={Kai Yi and Bingxin Zhou and Yiqing Shen and Pietro Lio and Yu Guang Wang},
booktitle={Thirty-seventh Conference on Neural Information Processing Systems},
year={2023},
url={https://openreview.net/forum?id=u4YXKKG5dX}
}