TranSHER
Codes for the paper TranSHER: Translating Knowledge Graph Embedding with Hyper-Ellipsoidal Restriction accepted by the EMNLP 2022.
Preparation
Environment Setup
Environment can be easily set up by the following commands:
# create python virtual environment, recommending using conda
conda create -n TranSHER python=3.8
conda activate TranSHER
conda install pytorch torchvision torchaudio pytorch-cuda=11.6 -c pytorch -c nvidia
conda install -c conda-forge scikit-learnOur codes are adapted from the RotatE and PairRE code bases, the environment requirements should be compatible.
Dataset
All the full-ranking datasets are contained in the repo [path to project]/data/. You can put provide your own customized dataset by adding processed data at the folder.
The OGB link prediction datasets can be referred to the OGB official repo.
The main difference between the full-ranking datasets and the OGB link prediction dataset is that
- the full-ranking datasets requires the model to retrieve matching entities from all the entity candidates, while
- the OGB datasets only require model to retrieve the correct candidate from a small subset, since the entity quantity could be up to 2,500K.
Experiments
Full-ranking Datasets
The TranSHER results on those "classic" full-ranking dataset can be reproduced by the following commands.
# FB15k-237
bash run.sh train TranSHER FB15k-237 0 0 1024 256 1500 6.0 1.0 0.00005 100000 16 -dr -r 0.00005
# DB100K
bash run.sh train TranSHER DB100K 0 0 1024 256 500 10.0 1.0 0.00005 150000 8 -dr -r 0.00001
# YAGO37
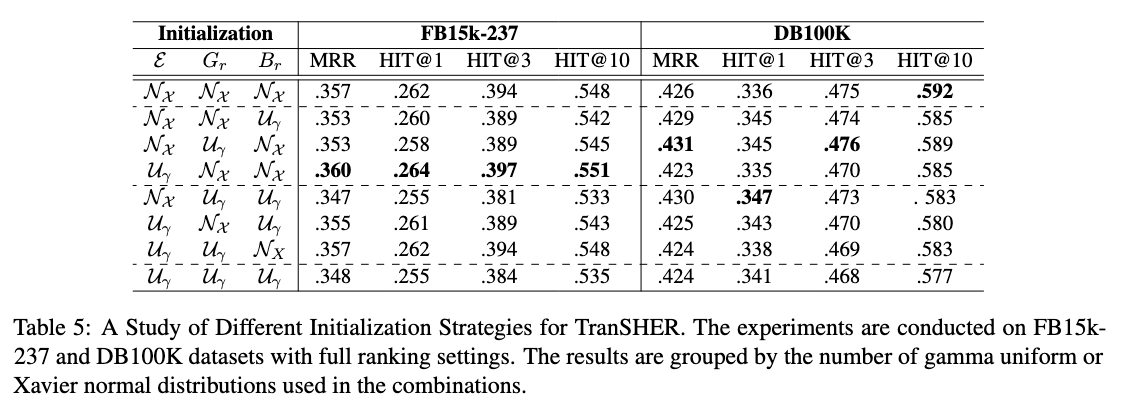
bash run.sh train TranSHER YAGO37 0 0 1024 256 500 14.0 1.0 0.00005 150000 8 -dr -r 0.00001The default initialization strategies are set as U_N_N' for TranSHER, i.e. uniform distribution for the entity embeddings and Xavier Normal for the relation and translation embeddings.
By adding parameters like '-ci U_N_N' to the end of shell script, you can modify the initialization strategy for different components of the model.
OGB Link Prediction
The experiments by adding TranSHER score function to the OGB codes in PairRE.
def TranSHER(self, head, relation, tail, mode):
re_head, re_tail, rel_bias = torch.chunk(relation, 3, dim=2)
head = F.normalize(head, 2, -1)
tail = F.normalize(tail, 2, -1)
score = head * re_head + rel_bias - tail * re_tail
score = self.gamma.item() - torch.norm(score, p=1, dim=2)
return scoreAnalysis Experiments
TBD
- relation-specific translation visualization
- standard variance of gradients during training
- initialization strategies
- case study
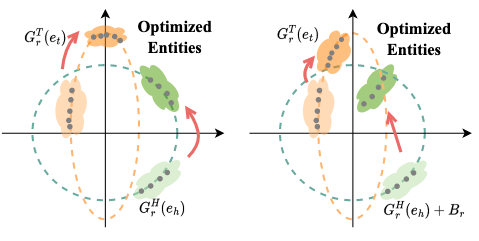
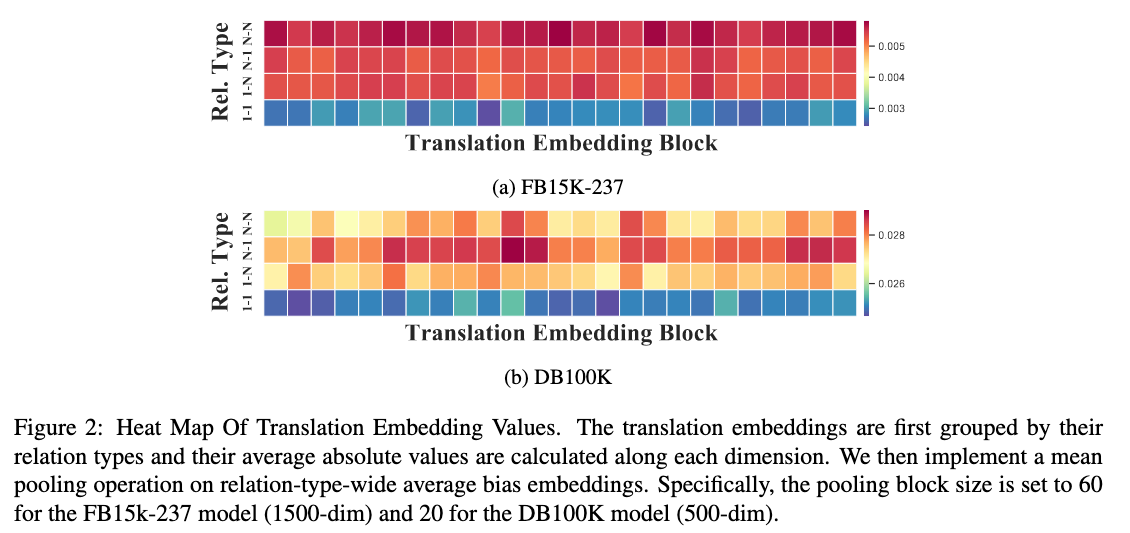
Relation-specific Translation Visualization
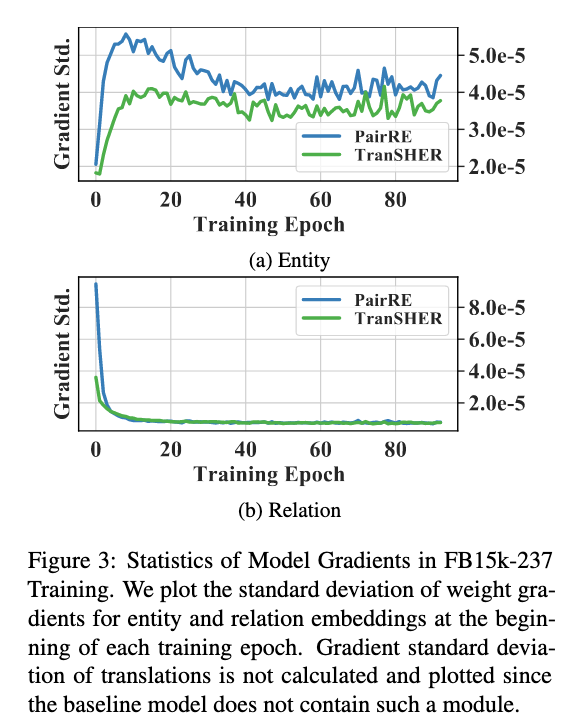
Standard Variance of Training Gradients
Initialization Strategies
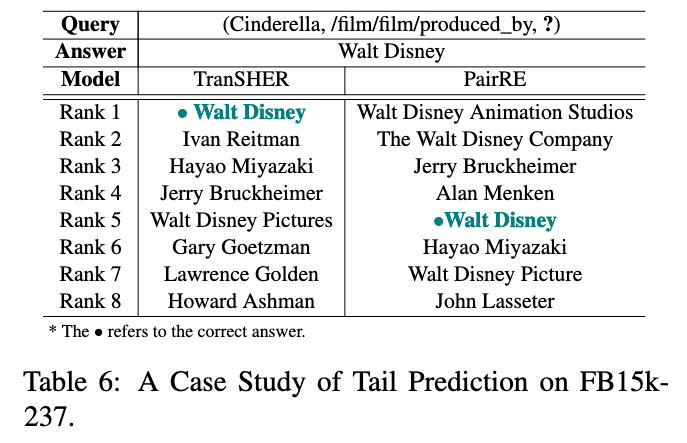
Case Study
Citation
For the arxiv version:
@article{li2022transher,
title={TransHER: Translating Knowledge Graph Embedding with Hyper-Ellipsoidal Restriction},
author={Li, Yizhi and Fan, Wei and Liu, Chao and Lin, Chenghua and Qian, Jiang},
journal={arXiv preprint arXiv:2204.13221},
year={2022}
}