Implementation for the paper: "RandStainNA: Learning Stain-Agnostic Features from Histology Slides by Bridging Stain Augmentation and Normalization" [MICCAI 2022]
Paper link: https://arxiv.org/abs/2206.12694
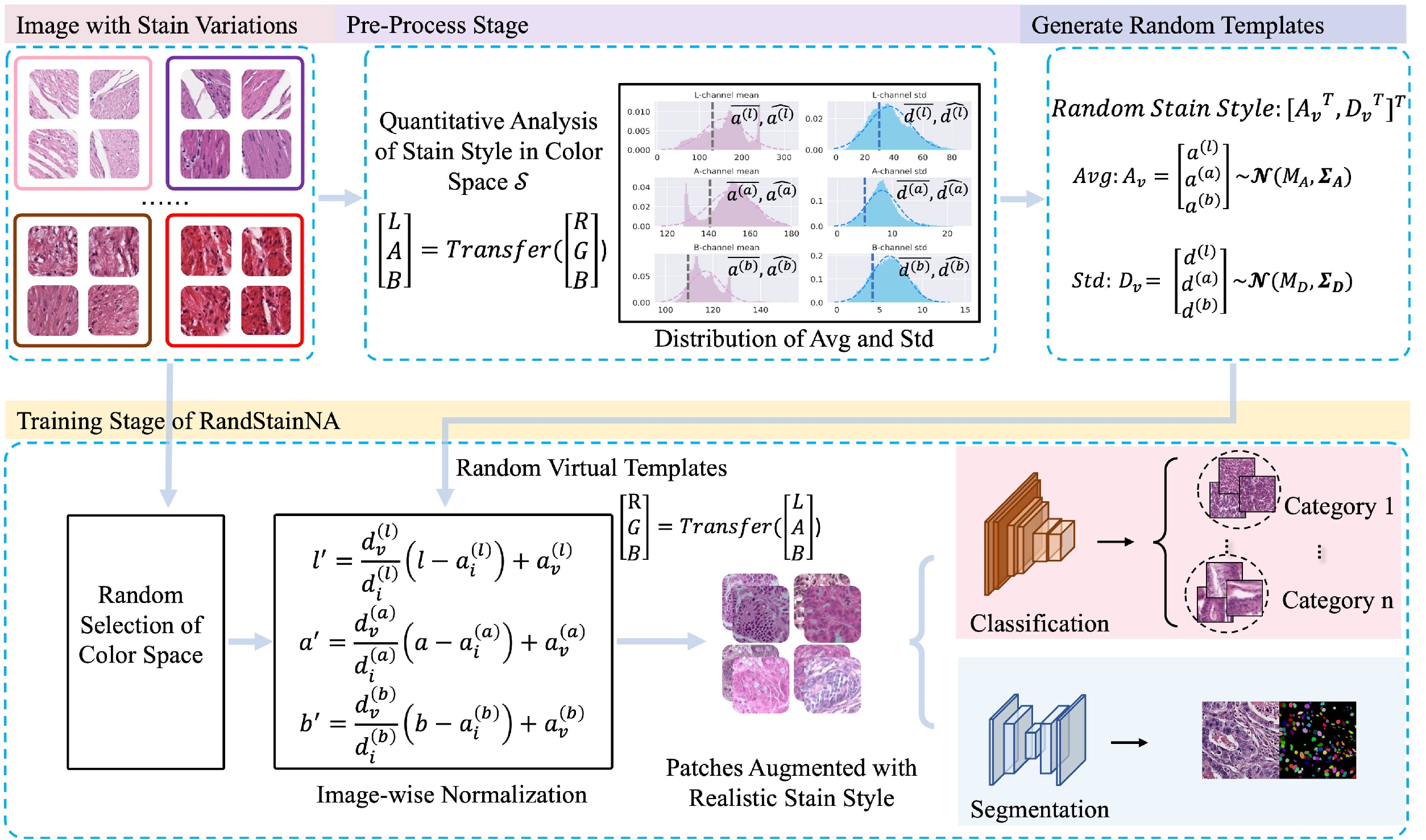
RandStainNA is a augmentation scheme to train a stain agnostic deep learning model specifically for histology analysis. It unifies Stain Normalization and Stain Augmentation by constraining variable stain styles in a practicable range. The RandStainNA is applicable to stain normalization in a collection of color spaces i.e., HED, HSV, LAB.
The codes are organized as follows:
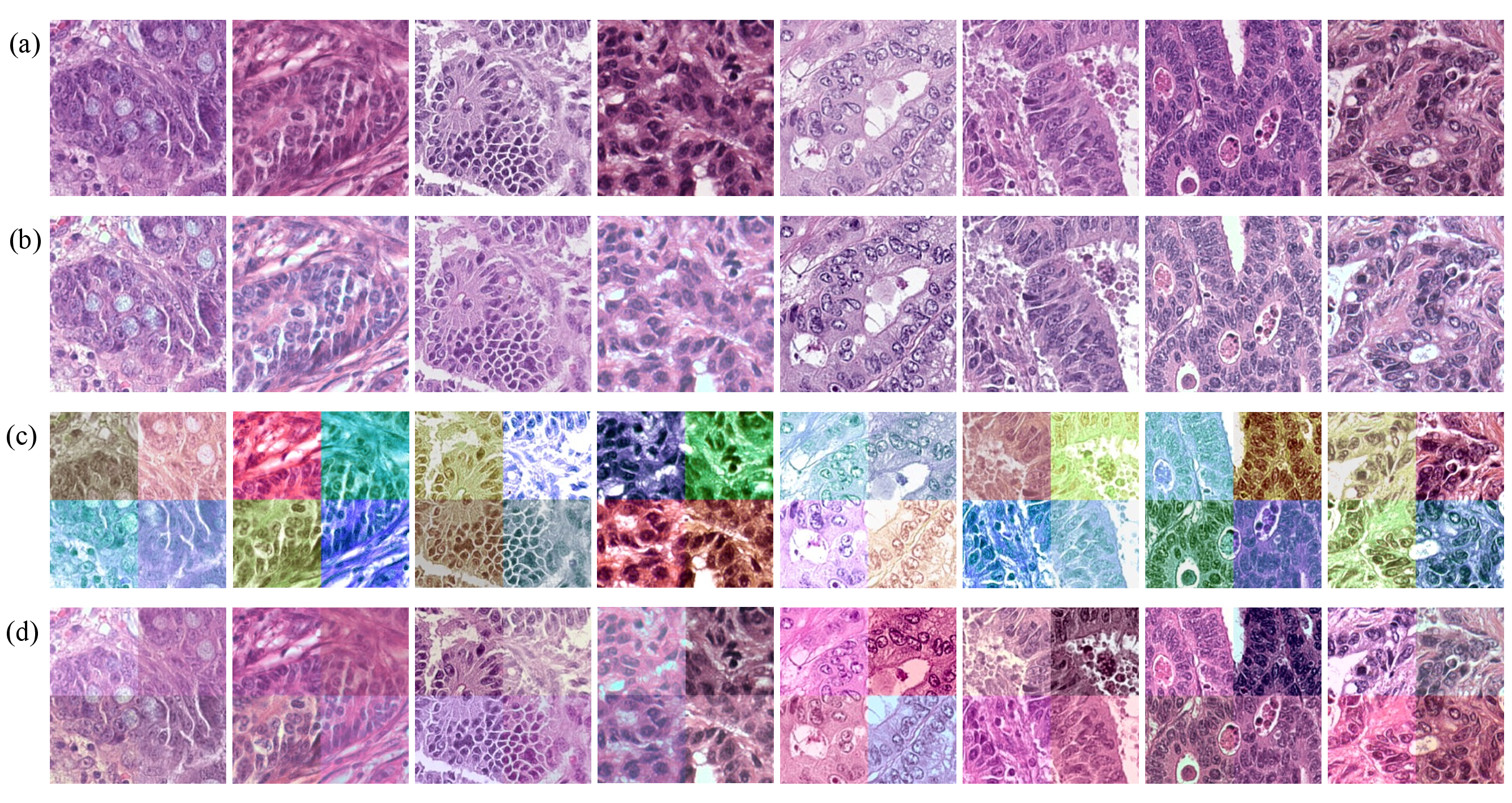
visualization: provides the visualization of stain normalizations, stain augmentations, and RandStainNA.origin: the raw images.randstainna: visualizations of images augmented by RandStainNA.stain_augmentation: visualizations of images augmented by stain augmentation.stain_augmentation: visualizations of images processed by stain normalization.
preprocess: perform the statistics in the pre-processing stage. Results should be presented asCRC_LAB_randomTrue_n0.yaml.randstainna.py: we warp RandStainNA in this file.main.py: provides the use case for RandStainNA inrandstainna.py.
Run
pip install -r requirements.txt
to install the required libraries. Note that opencv-python, pyyaml and scikit-image are necessary for RandStainNA; while we use fitter for performing the statistic analysis in the pre-processing stage.
Please refer to main.py for using RandStainNA:
transforms_list = [
RandStainNA(yaml_file='./CRC_LAB_randomTrue_n0.yaml', std_hyper=-0.3, probability=1.0,distribution='normal', is_train=True)
]
transforms.Compose(transforms_list)