- Introduction
- Download
- Tutorial
- QC Functions
- Imputation
- Data Visualization
- Utility Functions for WGCNA
- [WGCNA pipeline for SCD](#wgcna pipeline)
- Contact and feedback
- Citation
The VIQCing (Visualization, Imputation, Quality Control) package has been made to help processing Metabolomic Data. It contains a robust pipeline that can be adapted depending on the input type and it needs.
Source code can be downloaded from github.
git clone https://github.com/yilboudo/VIQCing.git
You can install in Rstudio using:
devtools::install_github("yilboudo/VIQCing") The data used as input usually is a LC-MS or NMR Feature Matrix, with the rows as metabolites and columns as samples. That said, as long as it looks like the following dataset, the data can be used with this package: (From the datasets::swiss)
## dummyCompound dummyMetabolite Courtelary Delemont Franches.Mnt
## 1 Fertility NA 80.20 83.10 92.5
## 2 Agriculture NA 17.00 45.10 39.7
## 3 Examination NA 15.00 6.00 5.0
## 4 Education NA 12.00 9.00 5.0
## 5 Catholic NA 9.96 84.84 93.4
## 6 Infant.Mortality NA 22.20 22.20 20.2
If you data contains NA values, you can filter the row with the
qualityControl() function.
Produces the following files:
1. “QC_data.txt”: Summary of the QC;
2. “REMOVED_QC_data.txt”: Summary of the removed metabolite;
3. output, cleaned dataset file(optional);
4. “REMOVED_output.txt”, the removed set of metabolite (optional)
It will also warn you about potential problems. (e.g. if a line has sd of 0 or NA) The function returns the cleaned dataset and QC summary.
#Write the data in a file
write.table(dat, file = "dummySet.txt",sep = "\t", row.names = FALSE)
result <- VIQCing::qualityControl("dummySet.txt", missing=0.2, compound=1, metabolite=2, sampleStart = 3)## [1] "Saving QC data"
result$dataset[,1:5]## Compound Metabolite Courtelary Delemont Franches.Mnt
## 1 Fertility NA 80.20 83.10 92.5
## 2 Agriculture NA 17.00 45.10 39.7
## 3 Examination NA 15.00 6.00 5.0
## 4 Education NA 12.00 9.00 5.0
## 5 Catholic NA 9.96 84.84 93.4
## 6 Infant.Mortality NA 22.20 22.20 20.2
result$QC## compound cohort metabolite nbna sd mu CV
## 1 Fertility NA 0 12.491697 70.14255 0.1780901
## 2 Agriculture NA 0 22.711218 50.65957 0.4483105
## 3 Examination NA 0 7.977883 16.48936 0.4838200
## 4 Education NA 0 9.615407 10.97872 0.8758220
## 5 Catholic NA 0 41.704850 41.14383 1.0136356
## 6 Infant.Mortality NA 0 2.912697 19.94255 0.1460544
## remove
## 1 FALSE
## 2 FALSE
## 3 FALSE
## 4 FALSE
## 5 FALSE
## 6 FALSE
If we had a line with only NA, another one with all the same value and a duplicate :
newDat <- result$dataset
newDat[nrow(newDat)+1,] <- 0
newDat[nrow(newDat)+1,] <- 1#Write the data in a file
write.table(newDat, file = "falsedummySet.txt",sep = "\t", row.names = FALSE)
result <- VIQCing::qualityControl("falsedummySet.txt", missing=1, compound=1, metabolite=2, sampleStart = 3)## [1] "Saving QC data"
## Warning in VIQCing::qualityControl("falsedummySet.txt", missing = 1,
## compound = 1, : row 8 is a duplicated compound_metabolite: NA_NA
## Warning in VIQCing::qualityControl("falsedummySet.txt", missing = 1,
## compound = 1, : Sd == NA for row: NA_NA
## Warning in VIQCing::qualityControl("falsedummySet.txt", missing = 1,
## compound = 1, : Sd == 0 for row: NA_NA
result$dataset[,1:5]## Compound Metabolite Courtelary Delemont Franches.Mnt
## 1 Fertility NA 80.20 83.10 92.5
## 2 Agriculture NA 17.00 45.10 39.7
## 3 Examination NA 15.00 6.00 5.0
## 4 Education NA 12.00 9.00 5.0
## 5 Catholic NA 9.96 84.84 93.4
## 6 Infant.Mortality NA 22.20 22.20 20.2
## 7 NA NA NA NA NA
## 8 NA NA 1.00 1.00 1.0
result$QC## compound cohort metabolite nbna sd mu CV
## 1 Fertility NA 0 12.491697 70.14255 0.1780901
## 2 Agriculture NA 0 22.711218 50.65957 0.4483105
## 3 Examination NA 0 7.977883 16.48936 0.4838200
## 4 Education NA 0 9.615407 10.97872 0.8758220
## 5 Catholic NA 0 41.704850 41.14383 1.0136356
## 6 Infant.Mortality NA 0 2.912697 19.94255 0.1460544
## 7 NA NA 47 NA NaN NA
## 8 NA NA 0 0.000000 1.00000 0.0000000
## remove
## 1 FALSE
## 2 FALSE
## 3 FALSE
## 4 FALSE
## 5 FALSE
## 6 FALSE
## 7 FALSE
## 8 FALSE
# Use the customisation output function:
VIQCing::QCcustomization(result$QC, REMOVE=FALSE)## compound cohort metabolite nbna sd mu CV
## 1 Fertility NA 0 12.491697 70.14255 0.1780901
## 2 Agriculture NA 0 22.711218 50.65957 0.4483105
## 3 Examination NA 0 7.977883 16.48936 0.4838200
## 4 Education NA 0 9.615407 10.97872 0.8758220
## 5 Catholic NA 0 41.704850 41.14383 1.0136356
## 6 Infant.Mortality NA 0 2.912697 19.94255 0.1460544
## 7 NA NA 47 NA NaN NA
## 8 NA NA 0 0.000000 1.00000 0.0000000
Impute the given dataset with different method options. Produces filename_imputed.txt, containing the imputed dataset;
Available imputation methods:
- “knn”: From the impute package, use the k nearest neighboors to impute the values;
- “RF”: From the missForest package, use RandomForest algorithm to impute the values;
- “QRILC”: From the imputeLCMD package, use Quantile regression to impute the values;
- “SVD”: From the pcaMethods package, use SVDimpute algorithm as proposed by Troyanskaya et al, 2001. to impute the values;
- “mean”,“median”, "“median”, “0”, “HM”: simple value replacement, either by the mean, median, 0 of Half minimum of the row;
returns the imputed Dataset
If we create some holes in the previous dataset:
## Compound Metabolite Courtelary Delemont Franches.Mnt
## 1 Fertility NA NA 83.10 92.5
## 2 Agriculture NA 17.00 45.10 39.7
## 3 Examination NA 15.00 6.00 NA
## 4 Education NA 12.00 9.00 5.0
## 5 Catholic NA 9.96 84.84 NA
## 6 Infant.Mortality NA NA 22.20 20.2
result <- VIQCing::imputation("holesdummySet.txt", method = "SVD", transformation = "scale", compound = 1, metabolite = 2, sampleStart = 3)## [1] "saving imputated data"
result[, 1:5]## Compound Metabolite Courtelary Delemont Franches.Mnt
## 1 Fertility NA 81.522 83.10 92.500
## 2 Agriculture NA 17.000 45.10 39.700
## 3 Examination NA 15.000 6.00 14.148
## 4 Education NA 12.000 9.00 5.000
## 5 Catholic NA 9.960 84.84 13.156
## 6 Infant.Mortality NA 26.609 22.20 20.200
You can use the NRMSE function to evaluate the accuracy of the imputation:
# Only input the Samples, not the compound/metabolite columns
VIQCing::NRMSE(result[,3:dim(result)[2]], dat[,3:dim(dat)[2]])## [1] 0.288641
You can also use the imputationTest() function, which will produce a more complete output of the NRMSE when asked. It is advised to test most of the methods and transformation on your own datasets to determine the optimal imputation method.
VIQCing::imputationTest("dummySet.txt", method="SVD", transformation = "scale", nbTest=15, sampleStart = 3)## [1] " Test run # 1"
## [1] " Test run # 2"
## [1] " Test run # 3"
## [1] " Test run # 4"
## [1] " Test run # 5"
## [1] " Test run # 6"
## [1] " Test run # 7"
## [1] " Test run # 8"
## [1] " Test run # 9"
## [1] " Test run # 10"
## [1] " Test run # 11"
## [1] " Test run # 12"
## [1] " Test run # 13"
## [1] " Test run # 14"
## [1] " Test run # 15"
## Method missing_proportion transformation NRMSE
## [1,] "SVD" "0.05" "scale" "0.123058948691335"
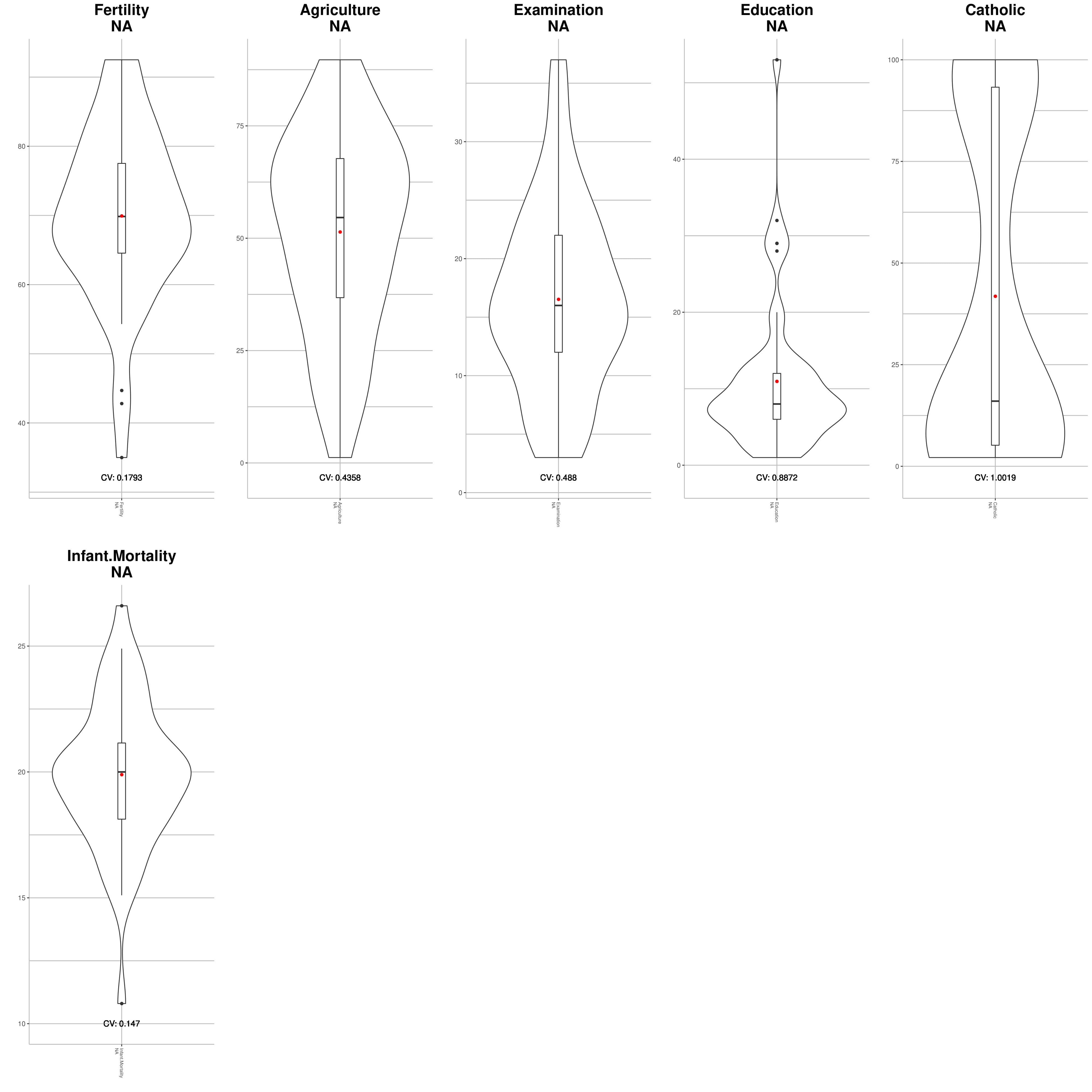
It is possible to visualize the distribution of your metabolomic data with Violin Plots. It will produce a .pdf file.
VIQCing::violinPlotQC("dummySet.txt", na=TRUE, compound=1, metabolite=2, sampleStart = 3)## [1] "Computing stats"
## [1] "Plotting"
## [1] "Saving PDF file"
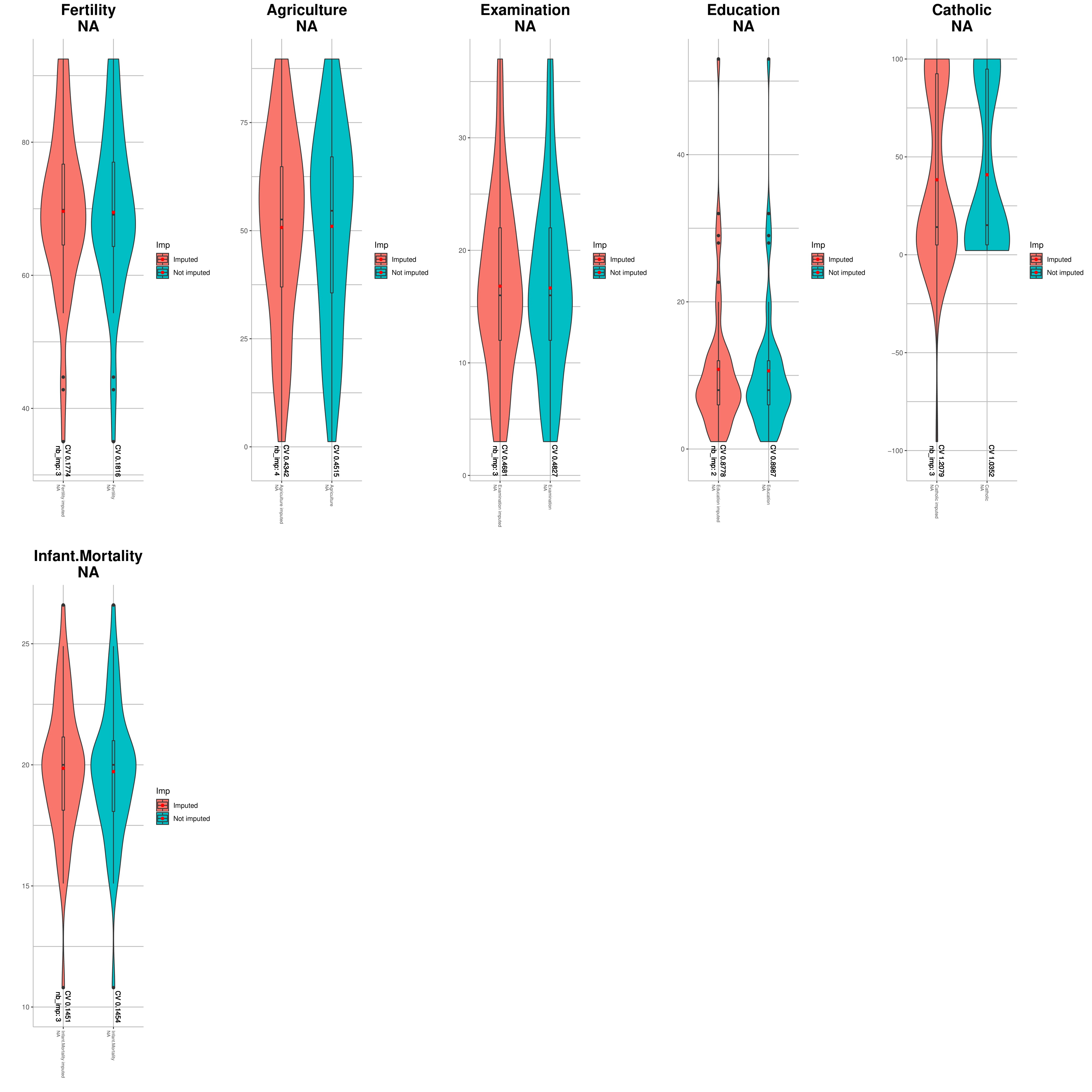
You can use violinPlotImp() to compare the distribution of your data before and after imputation:
VIQCing::violinPlotImp("holesdummySet.txt", "holesdummySet_imputed.txt",na=TRUE, compound=1, metabolite=2, sampleStart = 3, compoundImp = 1, metaboliteImp = 2, sampleStartImp = 3)## [1] "Computing stats"
## [1] "Plotting"
## [1] "Saving PDF file"
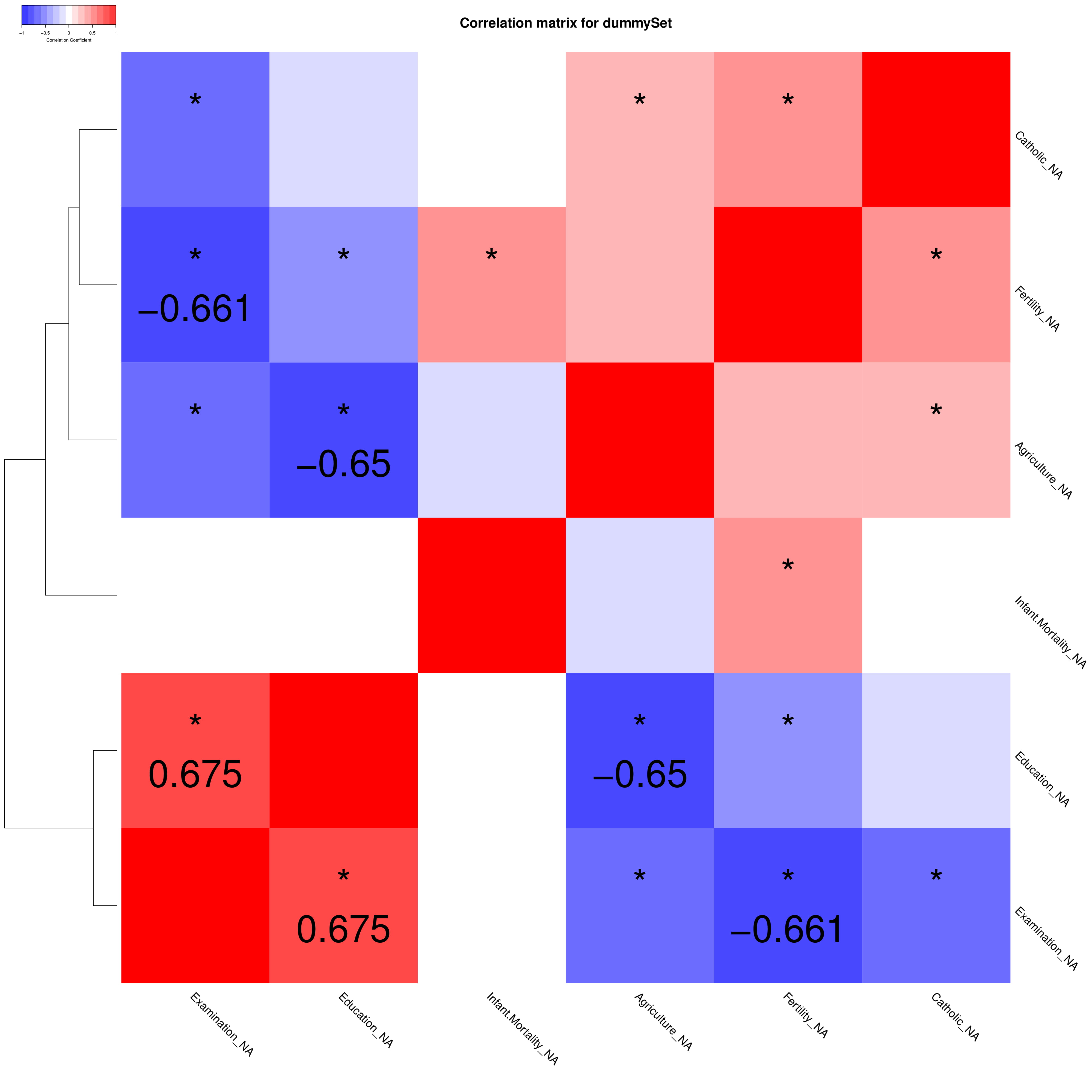
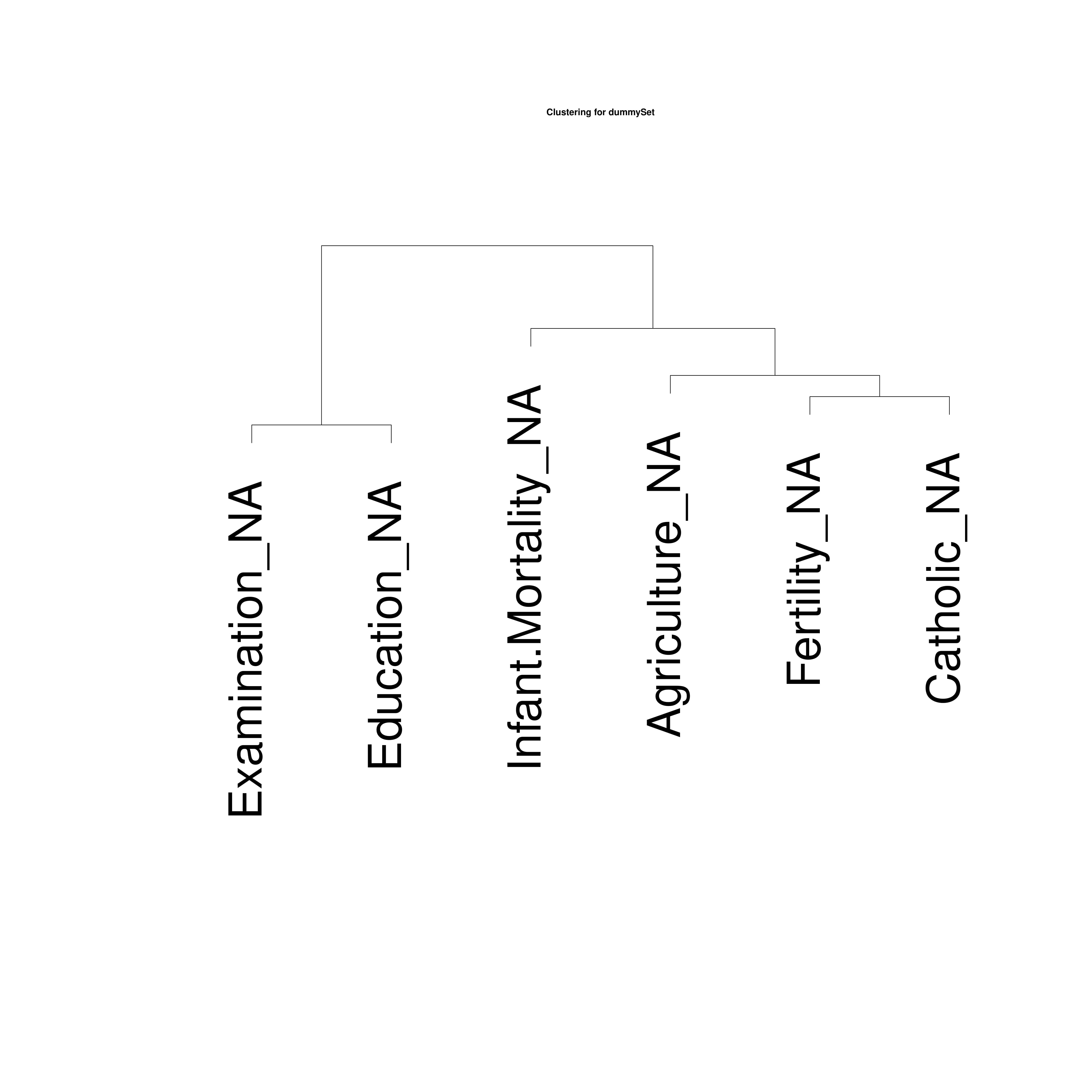
It is possible to build a correlation matrix and its associated
correlation tree for the given dataset. Both plots are optional and the
correlation test can be decided.
The function also produces:
1. “filename.pdf”, containing the asked plots;
2. “filname_pairs.txt”, containing the correlation pairs
returns the correlation matrix “r” and the p-value matrix
“P”
VIQCing::corMatrix("dummySet.txt", na=TRUE, compound=1, metabolite=2, sampleStart=3, testType="spearman", textSize = 5)## Warning in VIQCing::corMatrix("dummySet.txt", na = TRUE, compound = 1, metabolite = 2, : Application conditions for Spearman's Correlation test
## - Independent samples -> assumed
## Fertility_NA Agriculture_NA Examination_NA
## Fertility_NA 1.00 0.24 -0.66
## Agriculture_NA 0.24 1.00 -0.60
## Examination_NA -0.66 -0.60 1.00
## Education_NA -0.44 -0.65 0.67
## Catholic_NA 0.41 0.29 -0.48
## Infant.Mortality_NA 0.44 -0.15 -0.06
## Education_NA Catholic_NA Infant.Mortality_NA
## Fertility_NA -0.44 0.41 0.44
## Agriculture_NA -0.65 0.29 -0.15
## Examination_NA 0.67 -0.48 -0.06
## Education_NA 1.00 -0.14 -0.02
## Catholic_NA -0.14 1.00 0.07
## Infant.Mortality_NA -0.02 0.07 1.00
##
## n= 47
##
##
## P
## Fertility_NA Agriculture_NA Examination_NA
## Fertility_NA 0.1003 0.0000
## Agriculture_NA 0.1003 0.0000
## Examination_NA 0.0000 0.0000
## Education_NA 0.0018 0.0000 0.0000
## Catholic_NA 0.0039 0.0491 0.0007
## Infant.Mortality_NA 0.0021 0.3073 0.6929
## Education_NA Catholic_NA Infant.Mortality_NA
## Fertility_NA 0.0018 0.0039 0.0021
## Agriculture_NA 0.0000 0.0491 0.3073
## Examination_NA 0.0000 0.0007 0.6929
## Education_NA 0.3328 0.8992
## Catholic_NA 0.3328 0.6588
## Infant.Mortality_NA 0.8992 0.6588
Output file “dummySet_pairs.txt” :
## metabolite1 metabolite2 cor pvalue
## 1 Examination_NA Fertility_NA -0.6609030 4.281527e-07
## 2 Education_NA Agriculture_NA -0.6504638 7.457269e-07
## 3 Education_NA Examination_NA 0.6746038 1.998543e-07