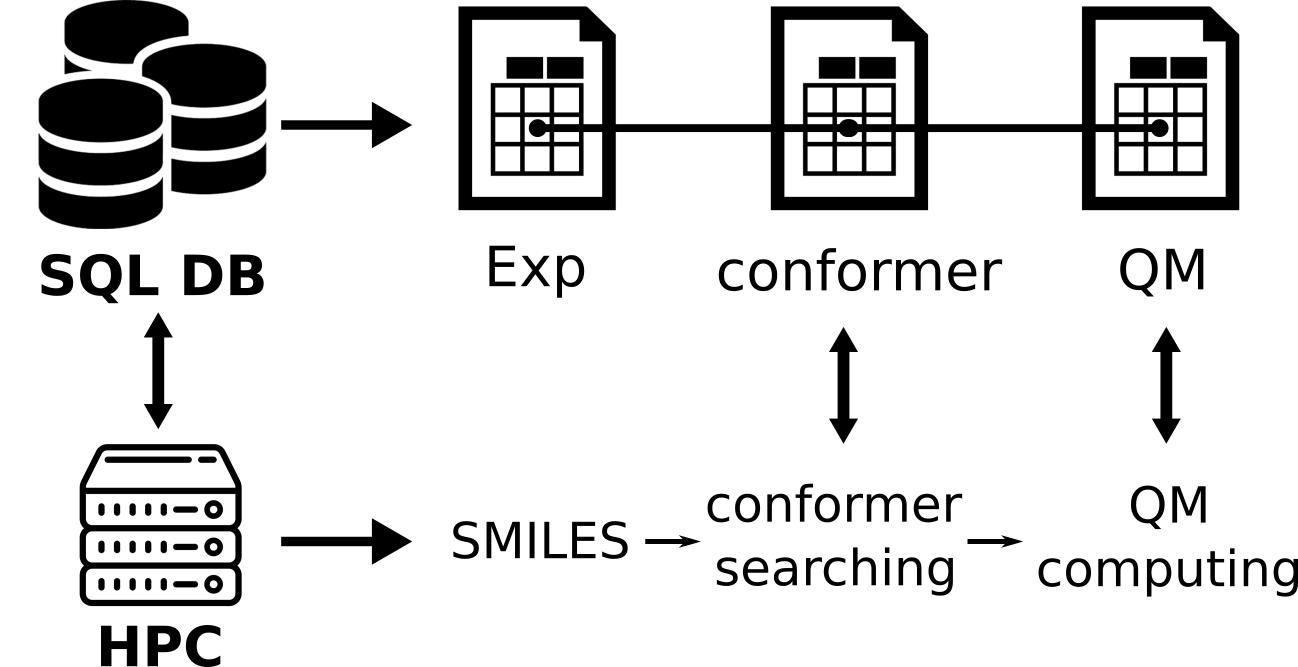
This repo contains code for high-throughput computations that feature a comprehensive conformer searching.
Dependencies are listed in the environment.yml.
Important software required are:
QM package Gaussian is required to run this workflow. Parameters for the Gaussian calculation need to be provided
in the config.py configure file. Users are responsible to setting the appropriate running environment for Gaussian.
For setting up the environment on a HPC see later section.
GFN2-xtb is used for geometry optimization. It can be either installed on your machine by the admin, or through a conda environment. For conda installation see later section.
The SQL type databse that holds all data. To run the workflow, a PostgreSQL service must be started, and the database
information must be provided in the config.py configure file, SQL parameters section.
To use the workflow on a HPC, you must have a Gaussian 09/16 installed on the HPC, and you are in the Gaussian group.
GFN2-xtb and PostgreSQL are strongly recommended to be installed on the cluster.
If the GFN-xtb is already installed, please specified the path to the executable xtb through the XTB_CMD parameter in config.py.
If the two are not installed,
you can still run the workflow by starting the PostgreSQL service through a Docker container or Conda environment;
and/or install the GFN-xtb through conda. In the following example, I will show how to run the workflow with only Conda.
Please skip this section if you have GFN-xtb and/or PostgreSQL installed on your HPC, or you are able to ask your admin to install these two for you.
GFN-xtb can be installed through Conda (see xtb repo). In that case, the xtb can be called directly through xtb, which is the default
option for XTB_CMD in config.py.
Please ref to this repo about starting a Postgres service in Conda environment, if you don't have PostgreSQL installed on the cluster. But note this is only a short-term solution. Talk to you admin about other options.
PS: If you start the Postgres service on the head node of a HPC, the computing node might not be able to see the service. This page might be helpful if you met this problem.
Three Postgres tables are related to this repo:
- exp_nmr stores the experimental data. This is not connected directly to the high-throughput QM workflow and is optional.
- compound stores the compound smiles, calculated QM properties and geometries, and status information. This table connects directly to the workflow and must be in the database. This table cannot be empty before running the workflow.
- conformers stores conformers information such as status. This table is populated throughout the workflow and can be empty at beginning.
The scripts/initialize_compound_db.py provides an example of initializing the three tables using the NMRShiftDB data.
To run the workflow, parameters including SQL database parameters, job details, conformer searching parameters, and Gaussian parameters
should be provided through the config.py file.
In principle, the workflow starts from a conformer searching under MMFF94 level, using RDKit.
A postpone RMSD and energy check is performed to remove high-energy and repeated conformers
(CONFORMER_E_THRESHOLD and CONFORMER_RMSD_THRESHOLD). Geometries of those conformers will be optimized through xtb semi-empirical method.
Each optimized geometry will be used to calculate QM properties (NMR chemical shift in this repo)
To run the workflow, simply do:
$ python main.py
A queueing software should be used if you run the workflow on a HPC. The submit_nmr.sh provides an example of submitting the calculation to the queue through Slurm.
You can write your own submitting bash script based on this example.
To use this workflow more efficiently, you can submit multiple bash script to run calculations in parallel. Calculations submitted through different bash scripts will act as separate workers, and will not affect each other.
The workflow is designed that requires minimum user intervention. We understand there is usually a wall time for each job
submitted to the HPC cluster. To save the computational resources and to make this workflow as efficient as possible, it would
be good to have the workflow resume from where the job was killed in the last run. To leverage this benefit, please specify the WALLTIME
parameter in the config.py. The workflow will then monitor the the remaining time of the job, and label the status as "timeout" towards end of the job.
Other running workers or next run will pick the "timeout" compound for calculation.
Due to the complexity of QM calculations and very different configurations of different HPCs, this workflow need to be carefully configured based on your HPC cluster.
On this repo, this workflow is implemented to run NMR chemical shift calculations though, in principle, you can easily revise this workflow for any QM properties including
molecular properties, atomic properties, and bond properties. For new properties, the worker/file_parser.py need to be revised to grab the corresponding
properties from the Gaussian output file.
This workflow is derived from Peter St. John's original workflow for BDE calculations: https://github.com/pstjohn/bde