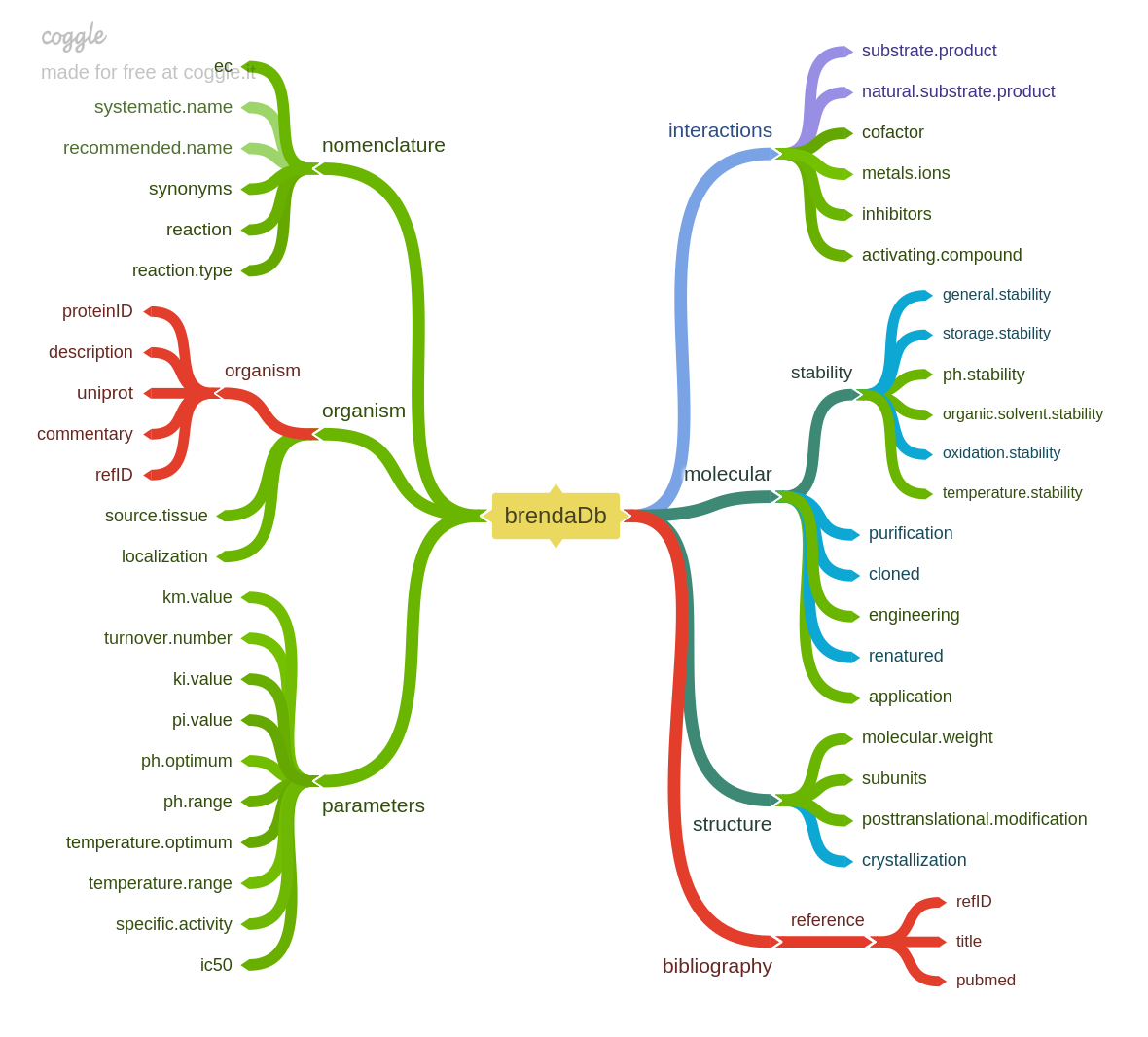
brendaDb aims to make importing and analyzing data from the BRENDA
database easier. The main functions
include:
- Read text file downloaded from
BRENDA
into an R
tibble - Retrieve information for specific enzymes
- Query enzymes using their synonyms, gene symbols, etc.
- Query enzyme information for specific BioCyc pathways
brendaDb is a Bioconductor package and can be installed through
BiocManager::install().
if (!requireNamespace("BiocManager", quietly = TRUE))
install.packages("BiocManager")
BiocManager::install("brendaDb", dependencies=TRUE)Alternatively, install the development version from GitHub.
if(!requireNamespace("brendaDb")) {
devtools::install_github("y1zhou/brendaDb")
}After the package is installed, it can be loaded into the R workspace by
library(brendaDb)To learn more about brendaDb, please refer to the vignette.
browseVignettes(package = "brendaDb")