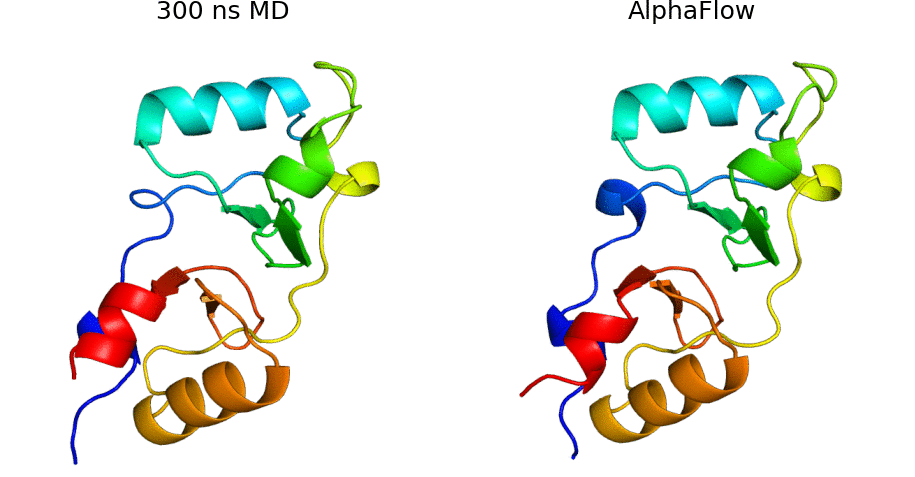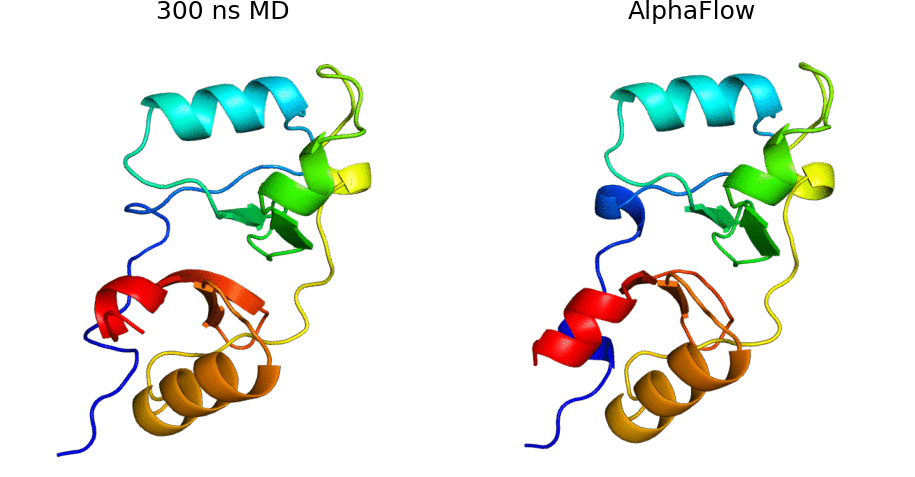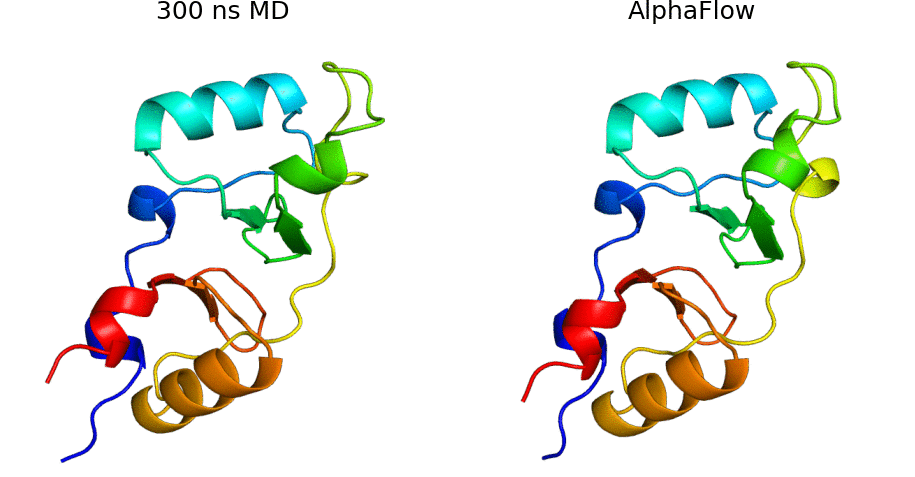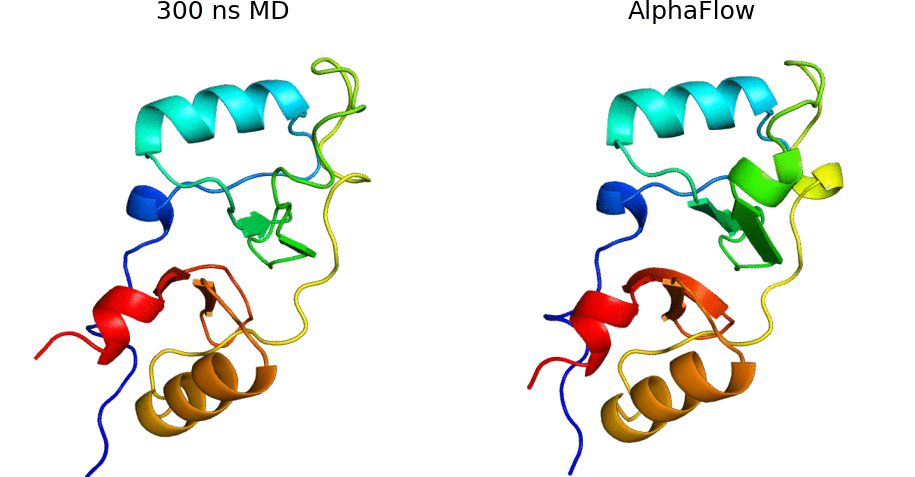
AlphaFlow is a modified version of AlphaFold, fine-tuned with a flow matching objective, capable of generative modeling of protein conformational ensembles. In particular, AlphaFlow models:
- Experimental ensembles, i.e, potential conformational states as they would be deposited in the PDB
- Molecular dynamics ensembles at physiological temperatures
We also provide a similarly fine-tuned version of ESMFold called ESMFlow. Technical details and thorough benchmarking results can be found in our paper, AlphaFold Meets Flow Matching for Generating Protein Ensembles, by Bowen Jing, Bonnie Berger, Tommi Jaakkola. This repository contains all code, instructions and model weights necessary to run the method. If you have any questions, feel free to open an issue or reach out at bjing@mit.edu.
In an environment with Python 3.9 (for example, conda create -n [NAME] python=3.9), run:
pip install numpy==1.21.2 pandas==1.5.3
pip install torch==1.12.1+cu113 -f https://download.pytorch.org/whl/torch_stable.html
pip install biopython==1.79 dm-tree==0.1.6 modelcif==0.7 ml-collections==0.1.0 scipy==1.7.1 absl-py einops
pip install pytorch_lightning==2.0.4 fair-esm mdtraj wandb
pip install 'openfold @ git+https://github.com/aqlaboratory/openfold.git@103d037'
We ran installation on a machine with CUDA 11.6 and have tested with A100 and A6000 GPUs.
We provide several versions of AlphaFlow (and similarly named versions of ESMFlow).
- AlphaFlow-PDB—trained on PDB structures to model experimental ensembles from X-ray crystallography or cryo-EM under different conditions
- AlphaFlow-MD—trained on all-atom, explicit solvent MD trajectories at 300K
- AlphaFlow-MD+Templates—trained to additionally take a PDB structure as input, and models the corresponding MD ensemble at 300K
For all models, the distilled version runs significantly faster at the cost of some loss of accuracy (benchmarked in the paper).
| Model | Version | Weights |
|---|---|---|
| AlphaFlow-PDB | base | https://alphaflow.s3.amazonaws.com/params/alphaflow_pdb_base_202402.pt |
| AlphaFlow-PDB | distilled | https://alphaflow.s3.amazonaws.com/params/alphaflow_pdb_distilled_202402.pt |
| AlphaFlow-MD | base | https://alphaflow.s3.amazonaws.com/params/alphaflow_md_base_202402.pt |
| AlphaFlow-MD | distilled | https://alphaflow.s3.amazonaws.com/params/alphaflow_md_distilled_202402.pt |
| AlphaFlow-MD+Templates | base | https://alphaflow.s3.amazonaws.com/params/alphaflow_md_templates_base_202402.pt |
| AlphaFlow-MD+Templates | distilled | https://alphaflow.s3.amazonaws.com/params/alphaflow_md_templates_distilled_202402.pt |
| Model | Version | Weights |
|---|---|---|
| ESMFlow-PDB | base | https://alphaflow.s3.amazonaws.com/params/esmflow_pdb_base_202402.pt |
| ESMFlow-PDB | distilled | https://alphaflow.s3.amazonaws.com/params/esmflow_pdb_distilled_202402.pt |
| ESMFlow-MD | base | https://alphaflow.s3.amazonaws.com/params/esmflow_md_base_202402.pt |
| ESMFlow-MD | distilled | https://alphaflow.s3.amazonaws.com/params/esmflow_md_distilled_202402.pt |
| ESMFlow-MD+Templates | base | https://alphaflow.s3.amazonaws.com/params/esmflow_md_templates_base_202402.pt |
| ESMFlow-MD+Templates | distilled | https://alphaflow.s3.amazonaws.com/params/esmflow_md_templates_distilled_202402.pt |
Training checkpoints (from which fine-tuning can be resumed) are available upon request; please reach out if you'd like to collaborate!
- Prepare a input CSV with an
nameandseqresentry for each row. Seesplits/atlas_test.csvfor examples. - If running an AlphaFlow model, prepare an MSA directory and place the alignments in
.a3mformat at the following paths:{alignment_dir}/{name}/a3m/{name}.a3m. If you don't have the MSAs, there are two ways to generate them:- Query the ColabFold server with
python -m scripts.mmseqs_query --split [PATH] --outdir [DIR]. - Download UniRef30 and ColabDB according to https://github.com/sokrypton/ColabFold/blob/main/setup_databases.sh and run
python -m scripts.mmseqs_search_helper --split [PATH] --db_dir [DIR] --outdir [DIR].
- Query the ColabFold server with
- If running an MD+Templates model, place the template PDB files into a templates directory with filenames matching the names in the input CSV. The PDB files should include only a single chain with no residue gaps.
The basic command for running inference with AlphaFlow is:
python predict.py --mode alphafold --input_csv [PATH] --msa_dir [DIR] --weights [PATH] --samples [N] --outpdb [DIR]
If running the PDB model, we recommend appending --self_cond --resample for improved performance.
The basic command for running inference with ESMFlow is
python predict.py --mode esmfold --input_csv [PATH] --weights [PATH] --samples [N] --outpdb [DIR]
Additional command line arguments for either model:
- Use the
--pdb_idargument to select (one or more) rows in the CSV. If no argument is specified, inference is run on all rows. - If running the MD model with templates, append
--templates_dir [DIR]. - If running any distilled model, append the arguments
--noisy_first --no_diffusion. - To truncate the inference process for increased precision and reduced diversity, append (for example)
--tmax 0.2 --steps 2. The default inference settings correspond to--tmax 1.0 --steps 10. See Appendix B.1 in the paper for more details.
To download and preprocess the PDB,
- Run
aws s3 sync --no-sign-request s3://pdbsnapshots/20230102/pub/pdb/data/structures/divided/mmCIF pdb_mmciffrom the desired directory. - Run
find pdb_mmcif -name '*.gz' | xargs gunzipto extract the MMCIF files. - From the repository root, run
python -m scripts.unpack_mmcif --mmcif_dir [DIR] --outdir [DIR] --num_workers [N]. This will preprocess all chains into NPZ files and create apdb_mmcif.csvindex. - Download OpenProteinSet with
aws s3 sync --no-sign-request s3://openfold/ openfoldfrom the desired directory. - Run
python -m scripts.add_msa_info --openfold_dir [DIR]to produce apdb_mmcif_msa.csvindex with OpenProteinSet MSA lookup. - Run
python -m scripts.cluster_chainsto produce apdb_clustersfile at 40% sequence similarity (Mmseqs installation required). - Create MSAs for the PDB validation split (
splits/cameo2022.csv) according to the instructions in the previous section.
To download and preprocess the ATLAS MD trajectory dataset,
- Run
bash scripts/download_atlas.shfrom the desired directory. - From the repository root, run
python -m scripts.prep_atlas --atlas_dir [DIR] --outdir [DIR] --num_workers [N]. This will preprocess the ATLAS trajectories into NPZ files. - Create MSAs for all entries in
splits/atlas.csvaccording to the instructions in the previous section.
Before running training, download the pretrained AlphaFold and ESMFold weights into the repository root via
wget https://storage.googleapis.com/alphafold/alphafold_params_2022-12-06.tar
tar -xvf alphafold_params_2022-12-06.tar params_model_1.npz
wget https://dl.fbaipublicfiles.com/fair-esm/models/esmfold_3B_v1.pt
The basic command for training AlphaFlow is
python train.py --lr 5e-4 --noise_prob 0.8 --accumulate_grad 8 --train_epoch_len 80000 --train_cutoff 2018-05-01 --filter_chains \
--train_data_dir [DIR] \
--train_msa_dir [DIR] \
--mmcif_dir [DIR] \
--val_msa_dir [DIR] \
--run_name [NAME] [--wandb]
where the PDB NPZ directory, the OpenProteinSet directory, the PDB mmCIF directory, and the validation MSA directory are specified. This training run produces the AlphaFlow-PDB base version. All other models are built off this checkpoint.
To continue training on ATLAS, run
python train.py --normal_validate --sample_train_confs --sample_val_confs --num_val_confs 100 --pdb_chains splits/atlas_train.csv --val_csv splits/atlas_val.csv --self_cond_prob 0.0 --noise_prob 0.9 --val_freq 10 --ckpt_freq 10 \
--train_data_dir [DIR] \
--train_msa_dir [DIR] \
--ckpt [PATH] \
--run_name [NAME] [--wandb]
where the ATLAS MSA and NPZ directories and AlphaFlow-PDB checkpoints are specified.
To instead train on ATLAS with templates, run with the additional arguments --first_as_template --extra_input --lr 1e-4 --restore_weights_only --extra_input_prob 1.0.
Distillation: to distill a model, append --distillation and supply the --ckpt [PATH] of the model to be distilled. For PDB training, we remove --accumulate_grad 8 and recommend distilling with a shorter --train_epoch_len 16000. Note that --self_cond_prob and --noise_prob will be ignored and can be omitted.
ESMFlow: run the same commands with --mode esmfold and --train_cutoff 2020-05-01.
We provide the ensembles sampled from the model which were used for the analyses and results reported in the paper.
| Model | Version | Samples |
|---|---|---|
| AlphaFlow-PDB | base | https://alphaflow.s3.amazonaws.com/samples/alphaflow_pdb_base_202402.zip |
| AlphaFlow-PDB | distilled | https://alphaflow.s3.amazonaws.com/samples/alphaflow_pdb_distilled_202402.zip |
| AlphaFlow-MD | base | https://alphaflow.s3.amazonaws.com/samples/alphaflow_md_base_202402.zip |
| AlphaFlow-MD | distilled | https://alphaflow.s3.amazonaws.com/samples/alphaflow_md_distilled_202402.zip |
| AlphaFlow-MD+Templates | base | https://alphaflow.s3.amazonaws.com/samples/alphaflow_md_templates_base_202402.zip |
| AlphaFlow-MD+Templates | distilled | https://alphaflow.s3.amazonaws.com/samples/alphaflow_md_templates_distilled_202402.zip |
| Model | Version | Samples |
|---|---|---|
| ESMFlow-PDB | base | https://alphaflow.s3.amazonaws.com/samples/esmflow_pdb_base_202402.zip |
| ESMFlow-PDB | distilled | https://alphaflow.s3.amazonaws.com/samples/esmflow_pdb_distilled_202402.zip |
| ESMFlow-MD | base | https://alphaflow.s3.amazonaws.com/samples/esmflow_md_base_202402.zip |
| ESMFlow-MD | distilled | https://alphaflow.s3.amazonaws.com/samples/esmflow_md_distilled_202402.zip |
| ESMFlow-MD+Templates | base | https://alphaflow.s3.amazonaws.com/samples/esmflow_md_templates_base_202402.zip |
| ESMFlow-MD+Templates | distilled | https://alphaflow.s3.amazonaws.com/samples/esmflow_md_templates_distilled_202402.zip |
MIT. Other licenses may apply to third-party source code noted in file headers.
@misc{jing2024alphafold,
title={AlphaFold Meets Flow Matching for Generating Protein Ensembles},
author={Bowen Jing and Bonnie Berger and Tommi Jaakkola},
year={2024},
eprint={2402.04845},
archivePrefix={arXiv},
primaryClass={q-bio.BM}
}