Variance-Aware Domain-Augmented Pseudo Labeling for Semi-Supervised Domain Generalization on Medical Image Segmentation
PyTorch implementation of Variance-Aware Domain-Augmented Pseudo Labeling for Semi-Supervised Domain Generalization on Medical Image Segmentation.
Huifeng Yao, Weihang Dai, Yiqun Lin, Xiaowei Hu, Xiaowei Xu, Xiaomeng Li
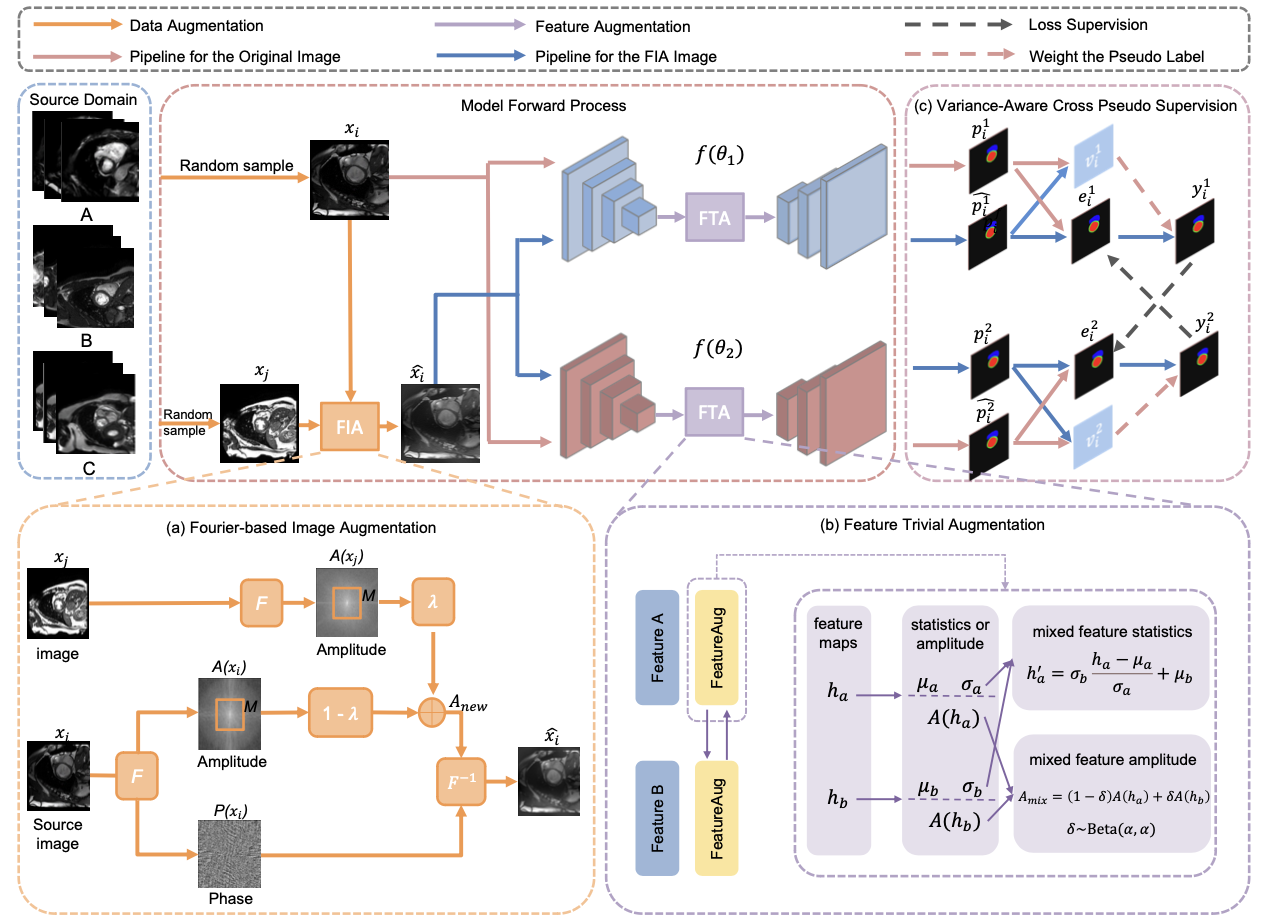
The overall framework
- We followed the setting of Semi-supervised Meta-learning with Disentanglement for Domain-generalised Medical Image Segmentation.
- We used two public datasets in this paper: Multi-Centre, Multi-Vendor & Multi-Disease Cardiac Image Segmentation Challenge (M&Ms) datast and Spinal cord grey matter segmentation challenge dataset
We followed the preprocessing of Semi-supervised Meta-learning with Disentanglement for Domain-generalised Medical Image Segmentation, you can find the preprocessing code here. After that, you should change the data directories in the dataloader(mms_dataloader or scgm_dataloader) file.
We use wandb to visulize our results. If you want to use this, you may need register an account first.
Use this command to install the environments.
conda env create -f VDPL_environment.yaml
If you want to train the model on M&Ms dataset, you can use this command. You can find the config information in config/mms.yaml.
bash mms_run.sh
If you want to run with multiple GPUs, you may need use accelerate config to set your environment.
If you want to evaluate our models on M&Ms dataset, you can use this command. And you should change the model name(line 201 and 202) and the test_vendor(line 199) to load different models.
python inference_mms.py