This is the official implementation of ICONIP 2023: Dynamic Data Augmentation via MCTS for Prostate MRI Segmentation.
To run the code base, first git clone the repo and install all requirements
pip install -r requirements.txtThen navigate into DDAug/. and execute
pip install -e . -
A Multi-site Dataset for Prostate MRI Segmentation (Subset 1-6)
- site A, B, C, D, E, F corresponds to subset 1, 2, 3, 4, 5, 6 respectively
-
nnUNet Prostate MRI dataset (Subset 7)
The folder structure follows the nnUNet folder structure. To ensure training runs without issue, you need environment variable nnUNet_raw_data_base, nnUNet_preprocessed, RESULTS_FOLDER ready. Expected folder structure is shown below.
-
Create folder for raw data and assign path to environment varialbe
nnUNet_raw_data_base, in which data are expected to follow:nnUNet_raw_data_base/ └── nnUNet_raw_data/ ├── Task001_Prostate_subset1/ │ ├── imagesTr/ │ │ ├── some_file_name_00_0000.nii.gz │ │ ├── some_file_name_01_0000.nii.gz │ │ ├── some_file_name_02_0000.nii.gz │ │ └── .... │ ├── imagesTs/ │ │ ├── some_file_name_03_0000.nii.gz │ │ ├── some_file_name_05_0000.nii.gz │ │ ├── some_file_name_11_0000.nii.gz │ │ └── ... │ ├── labelsTr/ │ │ ├── some_file_name_00.nii.gz │ │ ├── some_file_name_01.nii.gz │ │ ├── some_file_name_02.nii.gz │ │ └── .... │ └── dataset.json ├── Task002_Prostate_subset2/ │ ├── imagesTr/ │ │ └── ... │ ├── imagesTs/ │ │ └── ... │ ├── labelsTr/ │ │ └── ... │ └── dataset.jsonWhere the two digit
_00_insome_file_name_00_0000indicates scan number, and the four digit at the end indicates modality number. The essential content in dataset.json include:{ "name": "Prostate_RUNMC", "description": "", "reference": "", "licence": "", "release": "", "tensorImageSize": "4D", "modality": { "0": "MRI" }, "labels": { "0": "background", "1": "PZ", "2": "TZ" }, "numTraining": 30, "numTest": 0, "training": [ { "image": "./imagesTr/RUNMC_10.nii.gz", "label": "./labelsTr/RUNMC_10.nii.gz" }, { "image": "./imagesTr/RUNMC_06.nii.gz", "label": "./labelsTr/RUNMC_06.nii.gz" }, ... ], "test": [], "testing": [] }please note due to limited data size and as described in the paper, we reported mean DICE of 5-fold cross validation on the validation set using the weights of the last epoch.
-
After formatting all the raw data, create folder for processed data and assign to environment variable
nnUNet_preprocessed, then run the commandnnUNet_plan_and_preprocess -t Task001_Prostate_subset1
nnUNet will then create processed data in
nnUNet_preprocessed/ ├── Task001_Prostate_subset1/ │ └── .... ├── Task002_Prostate_subset2/ │ └── .... -
Finally you need to create folder for training result and assign to environment variable
RESULTS_FOLDER. Train logs, weights will be stored there.
To run 5-fold cross validation, make sure all environment variables are set, and execute
for fold in 0 1 2 3 4; do CUDA_VISIBLE_DEVICES=1 nnUNet_train 3d_fullres nnUNetTrainerV2_MCTS Task001_Prostate_subset1 $fold --npz; done;Once training completes, with the same environment variables, execute
python nnunet/inference/summarize_val_folds.pyThis will generate inference result and csv file with mean DICE score with weights using all 5-fold trainings. Please note the option --disable_tta is set to True in file nnunet/inference/summarize_val_folds.py.
You can use below code in jupyter notebook to have a nice visualization of all the results. (the for-else loop is not a bug)
result_folder = "RESULTS_FOLDER/nnUNet/3d_fullres"
for each_task in sorted(os.listdir(result_folder)):
print("-" * 100)
task_dir = f"{result_folder}/{each_task}"
for each_model in sorted(os.listdir(task_dir)):
model_dir = f"{task_dir}/{each_model}"
table = pd.DataFrame()
normal_exit = False
for fold in range(5):
each_fold = f"fold_{fold}"
if not os.path.isfile(f"{model_dir}/{each_fold}/testing/result.csv"):
continue
fold_result = pd.read_csv(f"{model_dir}/{each_fold}/testing/result.csv", index_col=0).drop(
index=["mean", "std"]
)
table = pd.concat([table, fold_result])
else:
normal_exit = True
# normal exit
mean_all = pd.DataFrame(
data=[table.mean(axis=0).to_numpy()], columns=table.columns, index=["mean"]
)
std_all = pd.DataFrame(data=[table.std(axis=0).to_numpy()], columns=table.columns, index=["std"])
table = pd.concat([table, mean_all, std_all])
table.to_csv(f"{model_dir}/fold_summary.csv")
print(f'out csv [{table.shape}] -> {each_task} {each_model.split("__")[0]}')
print(mean_all.to_string())
if not normal_exit:
print(f'failed with -> {each_task} {each_model.split("__")[0]}')
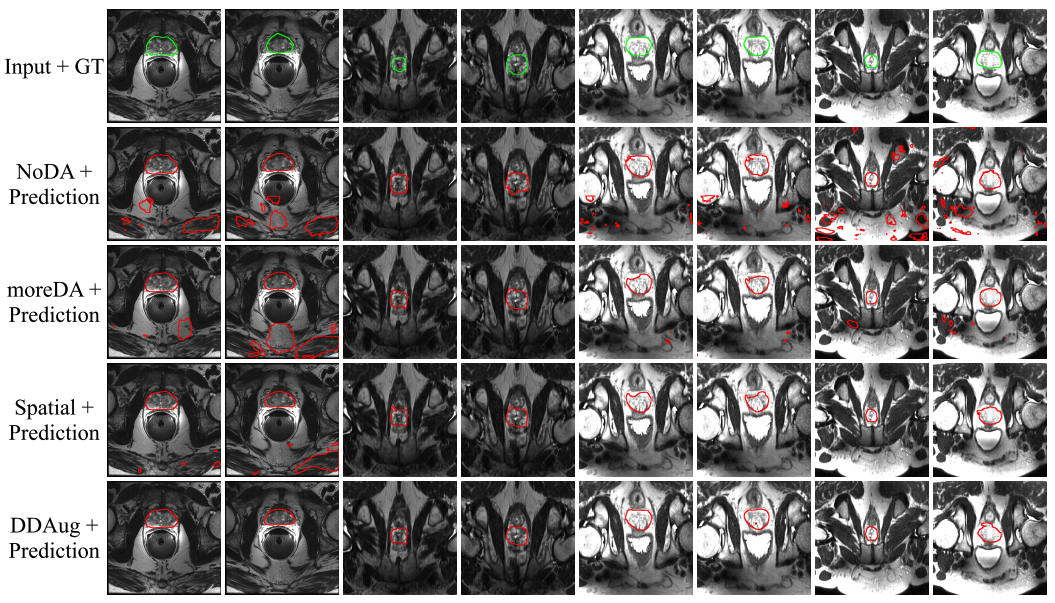
print("\n-----\n")You can use the draw_circle_using_mask.py to generate below images. Modify line 48, 60 and 61 for filename and folder paths.