DeepLabCut-live! SDK
This package contains a DeepLabCut inference pipeline for real-time applications that has minimal (software) dependencies. Thus, it is as easy to install as possible (in particular, on atypical systems like NVIDIA Jetson boards).
Performance: If you would like to see estimates on how your model should perform given different video sizes, neural network type, and hardware, please see: https://deeplabcut.github.io/DLC-inferencespeed-benchmark/
If you have different hardware, please consider submitting your results too! https://github.com/DeepLabCut/DLC-inferencespeed-benchmark
What this SDK provides: This package provides a DLCLive class which enables pose estimation online to provide feedback. This object loads and prepares a DeepLabCut network for inference, and will return the predicted pose for single images.
To perform processing on poses (such as predicting the future pose of an animal given it's current pose, or to trigger external hardware like send TTL pulses to a laser for optogenetic stimulation), this object takes in a Processor object. Processor objects must contain two methods: process and save.
- The
processmethod takes in a pose, performs some processing, and returns processed pose. - The
savemethod saves any valuable data created by or used by the processor
For more details and examples, see documentation here.
Note :: alone, this object does not record video or capture images from a camera. This must be done separately, i.e. see our DeepLabCut-live GUI.
News!
- March 2021: DeepLabCut-Live! version 1.0 is released, with support for tensorflow 1 and tensorflow 2!
- Feb 2021: DeepLabCut-Live! was featured in Nature Methods: "Real-time behavioral analysis"
- Jan 2021: full eLife paper is published: "Real-time, low-latency closed-loop feedback using markerless posture tracking"
- Dec 2020: we talked to RTS Suisse Radio about DLC-Live!: "Capture animal movements in real time"
Installation:
Please see our instruction manual to install on a Windows or Linux machine or on a NVIDIA Jetson Development Board. Note, this code works with tensorflow (TF) 1 or TF 2 models, but TF requires that whatever version you exported your model with, you must import with the same version (i.e., export with TF1.13, then use TF1.13 with DlC-Live; export with TF2.3, then use TF2.3 with DLC-live).
- available on pypi as:
pip install deeplabcut-live
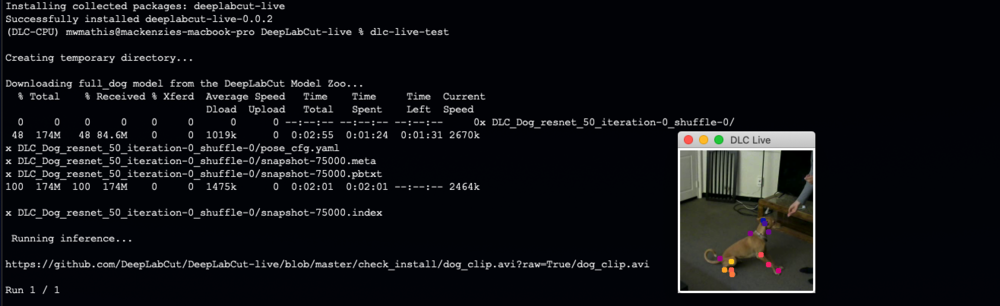
Note, you can then test your installation by running:
dlc-live-test
If installed properly, this script will i) create a temporary folder ii) download the full_dog model from the DeepLabCut Model Zoo, iii) download a short video clip of a dog, and iv) run inference while displaying keypoints. v) remove the temporary folder.
Quick Start: instructions for use:
- Initialize
Processor(if desired) - Initialize the
DLCLiveobject - Perform pose estimation!
from dlclive import DLCLive, Processor
dlc_proc = Processor()
dlc_live = DLCLive(<path to exported model directory>, processor=dlc_proc)
dlc_live.init_inference(<your image>)
dlc_live.get_pose(<your image>)DLCLive parameters:
path= string; full path to the exported DLC model directorymodel_type= string; the type of model to use for inference. Types include:base= the base DeepLabCut modeltensorrt= apply tensor-rt optimizations to modeltflite= use tensorflow lite inference (in progress...)
cropping= list of int, optional; cropping parameters in pixel number: [x1, x2, y1, y2]dynamic= tuple, optional; defines parameters for dynamic cropping of imagesindex 0= use dynamic cropping, boolindex 1= detection threshold, floatindex 2= margin (in pixels) around identified points, int
resize= float, optional; factor by which to resize image (resize=0.5 downsizes both width and height of image by half). Can be used to downsize large images for faster inferenceprocessor= dlc pose processor object, optionaldisplay= bool, optional; display processed image with DeepLabCut points? Can be used to troubleshoot cropping and resizing parameters, but is very slow
DLCLive inputs:
<path to exported model directory>= path to the folder that has the.pbfiles that you acquire after runningdeeplabcut.export_model<your image>= is a numpy array of each frame
Benchmarking/Analyzing your exported DeepLabCut models
DeepLabCut-live offers some analysis tools that allow users to peform the following operations on videos, from python or from the command line:
- Test inference speed across a range of image sizes, downsizing images by specifying the
resizeorpixelsparameter. Using thepixelsparameter will resize images to the desired number ofpixels, without changing the aspect ratio. Results will be saved (along with system info) to a pickle file if you specify an output directory.
python
dlclive.benchmark_videos('/path/to/exported/model', ['/path/to/video1', '/path/to/video2'], output='/path/to/output', resize=[1.0, 0.75, '0.5'])command line
dlc-live-benchmark /path/to/exported/model /path/to/video1 /path/to/video2 -o /path/to/output -r 1.0 0.75 0.5
- Display keypoints to visually inspect the accuracy of exported models on different image sizes (note, this is slow and only for testing purposes):
python
dlclive.benchmark_videos('/path/to/exported/model', '/path/to/video', resize=0.5, display=True, pcutoff=0.5, display_radius=4, cmap='bmy')command line
dlc-live-benchmark /path/to/exported/model /path/to/video -r 0.5 --display --pcutoff 0.5 --display-radius 4 --cmap bmy
- Analyze and create a labeled video using the exported model and desired resize parameters. This option functions similar to
deeplabcut.benchmark_videosanddeeplabcut.create_labeled_video(note, this is slow and only for testing purposes).
python
dlclive.benchmark_videos('/path/to/exported/model', '/path/to/video', resize=[1.0, 0.75, 0.5], pcutoff=0.5, display_radius=4, cmap='bmy', save_poses=True, save_video=True)command line
dlc-live-benchmark /path/to/exported/model /path/to/video -r 0.5 --pcutoff 0.5 --display-radius 4 --cmap bmy --save-poses --save-video
License:
This project is licensed under the GNU AGPLv3. Note that the software is provided "as is", without warranty of any kind, express or implied. If you use the code or data, we ask that you please cite us! This software is available for licensing via the EPFL Technology Transfer Office (https://tto.epfl.ch/, info.tto@epfl.ch).
Community Support, Developers, & Help:
This is an actively developed package and we welcome community development and involvement.
-
If you want to contribute to the code, please read our guide here, which is provided at the main repository of DeepLabCut.
-
We are a community partner on the
. Please post help and support questions on the forum with the tag DeepLabCut. Check out their mission statement Scientific Community Image Forum: A discussion forum for scientific image software.
-
If you encounter a previously unreported bug/code issue, please post here (we encourage you to search issues first): https://github.com/DeepLabCut/DeepLabCut-live/issues
Reference:
If you utilize our tool, please cite Kane et al, eLife 2020. The preprint is available here: https://www.biorxiv.org/content/10.1101/2020.08.04.236422v2
@Article{Kane2020dlclive,
author = {Kane, Gary and Lopes, Gonçalo and Sanders, Jonny and Mathis, Alexander and Mathis, Mackenzie},
title = {Real-time, low-latency closed-loop feedback using markerless posture tracking},
journal = {eLife},
year = {2020},
}