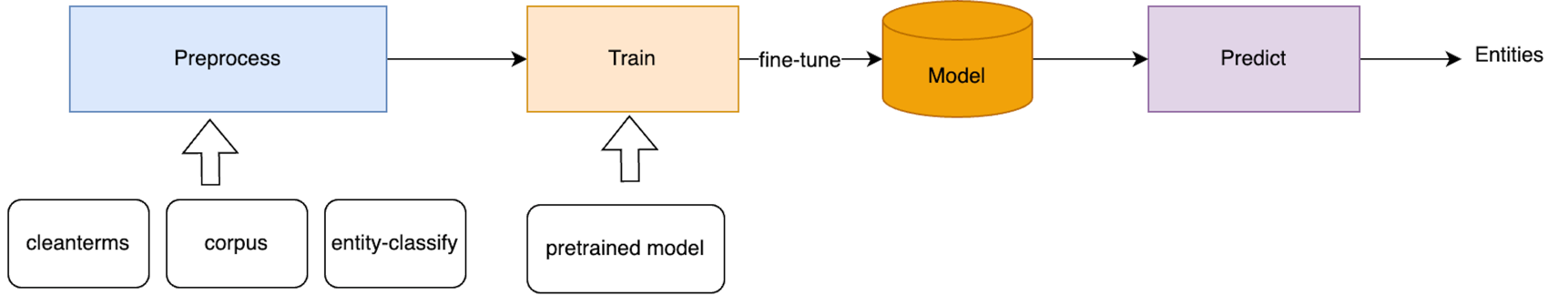
 The NER model employed in BIOS is the standard BERT sequential classifier.
Our approach of using automated annotation to generate training data belongs to the distant supervision (DS-NER) category.
The NER model employed in BIOS is the standard BERT sequential classifier.
Our approach of using automated annotation to generate training data belongs to the distant supervision (DS-NER) category.
- pretrain: scripts for downloading pretrained models
- example: small dataset to demo the code
- preprocess: prepare training data using automated annotation
- train: train codes
- predict: predict codes
- utils: utils codes
For optimal performance, we recommend a computer with following specs:
RAM: 16+ GB
CPU: 4+ cores, 3.3+ GHz/core
GPU Memory: 40 GB
The package is tested on Linux 20.04 operating system.
Python3.6 is recommended! For python3.7, see issues 14559.
!pip install -r requirements.txt
- Download cleanterms5.txt (password: d9ol) and put it under
./example/cleanterms/. - Download pretrained models:
cd pretrain && sh download_pubmedbert.sh
which will take about a few minutes to complete the download.
Prepare train/dev/test datasets.
Make sure cleanterms5.txt has been downloaded, and then:
1. cd preprocess
2. bash run.sh
Expect result: train/dev/test.txt under ../example/data/ner_train
Expect time: about 2-3 minutes.
Make sure pretrained model has been downloaded, and then:
1. cd train
2. sh train.sh
Expect result: fine-tuned model under ../example/data/ner_train/output
Expect time cost: about 5 minutes.
The evaluation result may be not good due to small training datasets.
configure fine-tuned model path (password: igg3) in the predict.sh, and then:
1. cd predict
2. bash predict.sh
Expect result: predictions for NER under ../example/data/ner_predict
Expect time cost: about 1 minute.
-
generate your cleanterms.txt from UMLS by your rules.
-
prepare your training texts and the STA results using STA model.
-
use your datasets to train and predict!
@misc{https://doi.org/10.48550/arxiv.2203.09975,
doi = {10.48550/ARXIV.2203.09975},
url = {https://arxiv.org/abs/2203.09975},
author = {Yu, Sheng and Yuan, Zheng and Xia, Jun and Luo, Shengxuan and Ying, Huaiyuan and Zeng, Sihang and Ren, Jingyi and Yuan, Hongyi and Zhao, Zhengyun and Lin, Yucong and Lu, Keming and Wang, Jing and Xie, Yutao and Shum, Heung-Yeung},
keywords = {Computation and Language (cs.CL), Machine Learning (cs.LG), FOS: Computer and information sciences, FOS: Computer and information sciences},
title = {BIOS: An Algorithmically Generated Biomedical Knowledge Graph},
publisher = {arXiv},
year = {2022},
copyright = {Creative Commons Attribution Non Commercial Share Alike 4.0 International}
}