This project was developed and tested for Ubuntu and Windows
To use the provided tools, the first step is to clone this repository, which can be can be accomplished by:
user@computer:~$ git clone https://github.com/vanluwin/proj-pca.gitThis section is a guide to the installations of a python environment with the requirements of this repository.
First, install Anaconda or Miniconda, both of them give you similar results, but the latter requires less disk space.
Now, create a python virtual environment and install the required packages following the commands. Substitute <environment_name> with a name for your environment
Open your terminal and execute the following commands:
user@computer:~$ conda create -n <enviroment_name> anaconda python=3
user@computer:~$ conda activate <enviroment_name> || source activate <enviroment_name>
(<enviroment_name>) user@computer:~$ conda install -c loopbio -c conda-forge -c pkgw-forge ffmpeg gtk2 numpy==1.16.3 opencv==3.4.3 matplotlib scipy pyserialOpen your anaconda command prompt and execute the following commands:
C:\Users\your-user> conda create -n <enviroment_name> anaconda python=3
C:\Users\your-user> conda activate <enviroment_name> || source activate <enviroment_name>
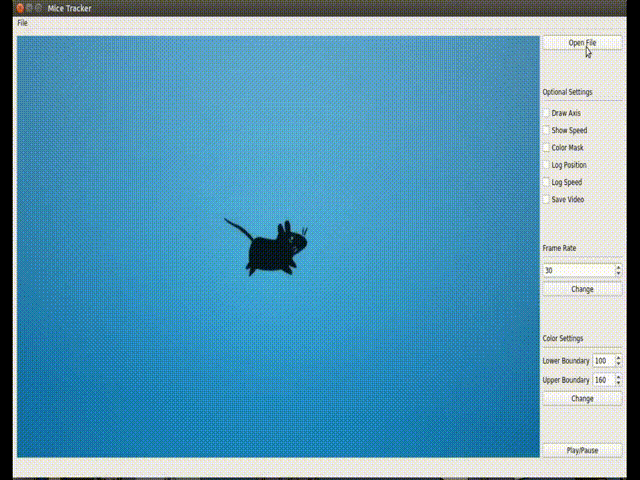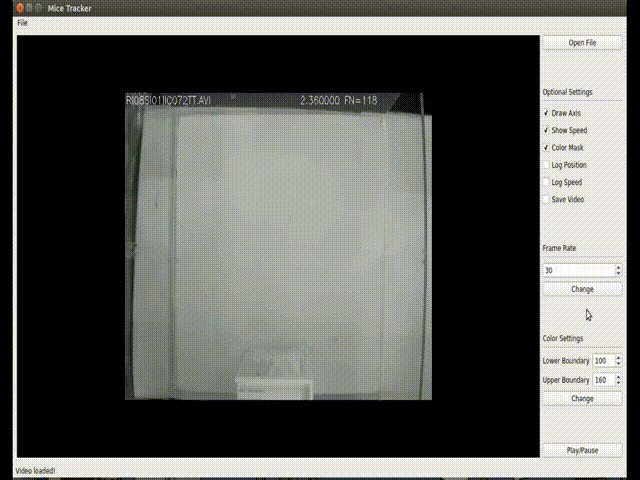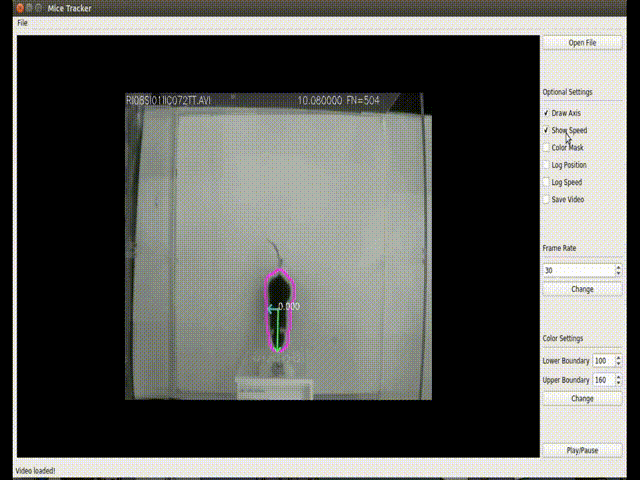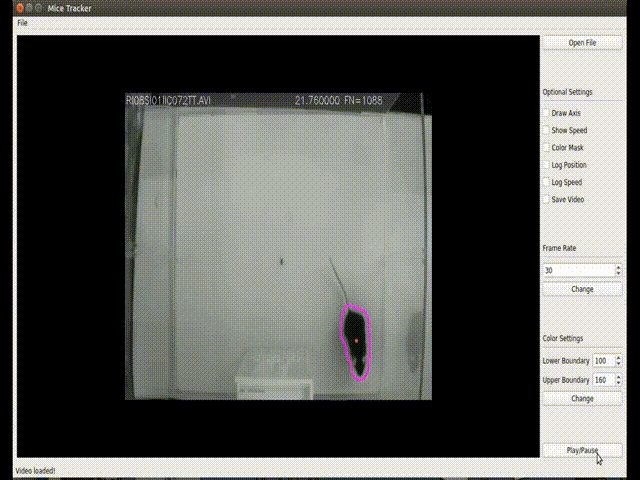
(<enviroment_name>) C:\Users\your-user> conda install -c loopbio -c conda-forge -c pkgw-forge ffmpeg numpy==1.16.3 opencv==3.4.3 matplotlib scipy pyserialThis GUI implements a more user-friendly way to interact with the tracking algorithm. Usage:
(<enviroment_name>) user@computer:~/proj-pca/gui$ python main.pyTo use the provided scripts, first make sure to activate your python environment:
On your terminal, execute the following command:
user@computer:~$ conda activate <enviroment_name>On your anaconda command prompt, execute the following command:
C:\Users\your-user> conda activate <enviroment_name>This script aims to track the mice and detect the head direction during behavioral neuroscience experiments. For testing, use the following suggested commands:
(<enviroment_name>) user@computer:~/proj-pca$ python pcaAnalyser.py [-h] [--color-mask] [--both-axis] [--show-mask] [--save-video] videoRequired arguments:
- video: Path to the video file to be processed.
Optional arguments:
- -h, --help: Show a help message and exit.
- --color-mask: Draw a colored mask over the detection.
- --both-axis: Draw both PCA axis.
- --show-mask: Displays a window with the segmented mask.
- --save-video: Create a video file with the analysis result.
This script aims to track mice throughout a neuroscience experiment detecting when the mice are present in a previously selected region, with that the program is able to keep track of how many frames the animal stayed inside each zone. Usage:
(<enviroment_name>) user@computer:~/proj-pca$ python tracker.py video frame_rate [--draw-axis] [--save-video] [--color-mask] [--log-position] [--log-speed]Required arguments:
- video: Path to the video file to be processed.
- frame rate: Frame rate of the video file to be processed.
Optional arguments:
- -h, --help: Shows a help message and exit.
- --draw-axis: Draws both axis found through PCA.
- --color-mask: Draws a colored mask over the detection.
- --save-video: Creates a video file with the analysis results.
- --log-position: Creates a log file with the (x, y) position coordinates of the tracked animal.
- --log-speed: Creates a log file with the speed of the tracked animal.
A statistics file containing the following information will be created.
Counters for the regions considering 30fps video
Traveled distance: 16971.568 pixels
Region 0: 3995 frames, 133.167s
Region 1: 4105 frames, 136.833s
Region 2: 852 frames, 28.400s
Region 3: 727 frames, 24.233s
Region 4: 1378 frames, 45.933s
This script is an integration of the tracking system with Arduino-based development boards, enabling experiments with real-time decisions like the control of a laser simulated by the blue led in the gif above.
To utilize the script first, upload the Arduino file to your microcontroller and make sure that the serial communication is working. Then, in your terminal, run the following command:
(<enviroment_name>) user@computer:~/proj-pca$ python trackerArduino.py [-h] [--draw-axis] [--save-video] [--color-mask] [--log-position] videoRequired arguments:
- video: Path to the video file to be processed.
Optional arguments:
- -h, --help: Show a help message and exit.
- --draw-axis: Draw both PCA axis.
- --color-mask: Draw a colored mask over the detection.
- --save-video: Create a video file with the analysis results.
- --log-position: Creates a text file with the (x, y) position of the tracked mice.
This script is intended to manually correct errors in detections that have already been made and can be edited, a window containing the instructions will be displayed. Pause and press d to edit the detections of the current frame. Usage:
(<enviroment_name>) user@computer:~/proj-pca$ python detectionsAnalyser.py video log_fileAvalible commands:
- space - Pause the video stream
- q, esc - Finish the execution
- s - Increases the delay between each video frame
- f - Decreases the delay between each video frame
- d - Opens a new window where the user is able to select a new point by drawing a rectangle with the disered point in its cente, to finish the selection press enter, press c to clean the selection.
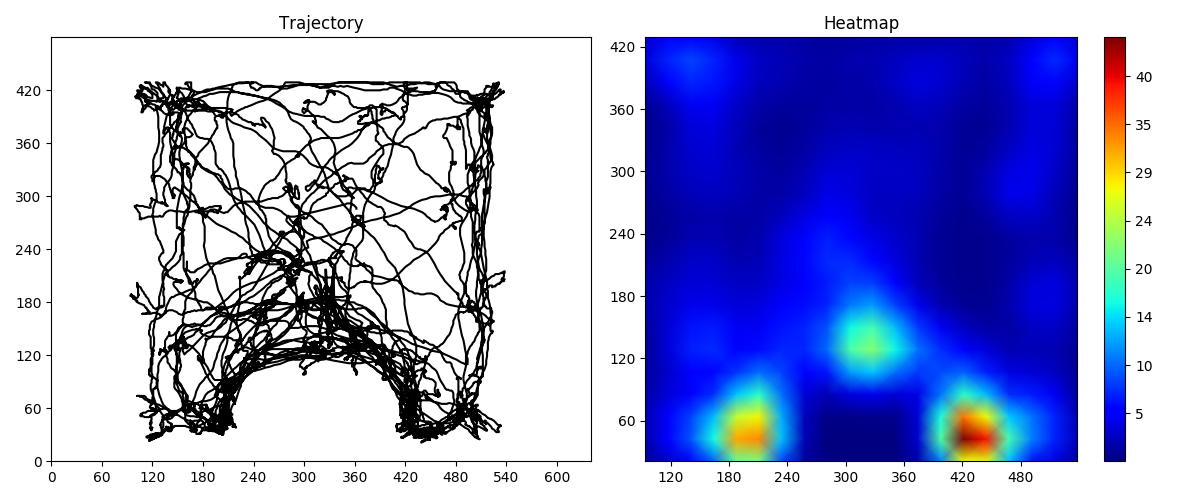
This scripts takes as input a detection log file produced by the tracker.py script used with the --log-position option, and produces a heatmap plot, a window containg the plot will be displayed. Usage:
(<enviroment_name>) user@computer:~/proj-pca$ python heatmapPlot.py log_file frameWidth frameHeightRequired arguments:
- log_file: Path to the log file file to be processed.
- frameWidth: Frame width of the processed video.
- frameHeight: Frame height of the processed video.
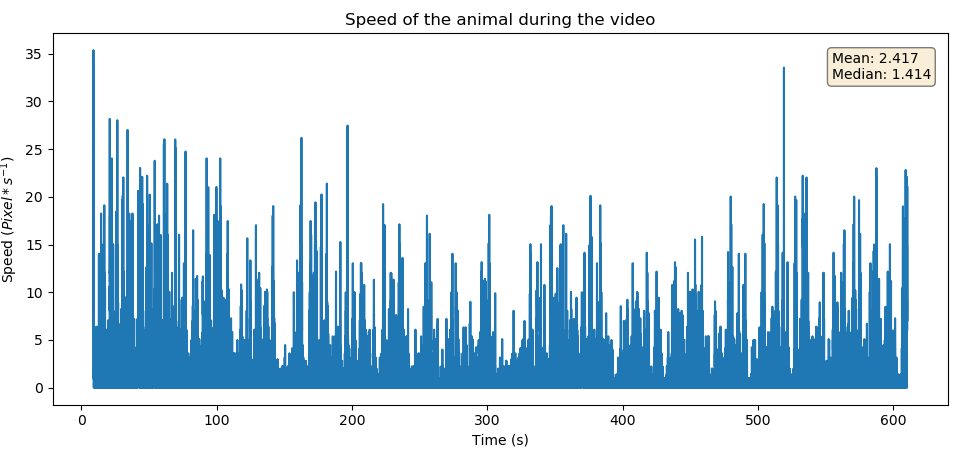
This scripts takes as input a speed log file produced by the tracker.py script used with the --log-speed option, and produces a speed plot, a window containg the plot will be displayed. Usage:
(<enviroment_name>) user@computer:~/proj-pca$ python speedPlot.py log_fileRequired arguments:
- log_file: Path to the log file file to be processed.