This repository contains codes for two papers:
- "Improved Brain Age Estimation with Slice-based Set Networks," ISBI 2021 [Link]. To find the code released at the time of this paper, see v1 release. Nevertheless, we recommend using the current version, which may produce slightly different results.
- "Transferring Models Trained on Natural Images to 3D MRI via Position Encoded Slice Models," ISBI 2023 [Link]
To cite the papers, please use the following BibTeX:
@inproceedings{gupta2023transferring,
title={Transferring Models Trained on Natural Images to 3D MRI via Position Encoded Slice Models},
author = {Gupta, Umang and Chattopadhyay, Tamoghna and Dhinagar, Nikhil and Thompson, Paul M. and Steeg, Greg Ver and Initiative, The Alzheimer's Disease Neuroimaging},
booktitle={2023 IEEE 20th International Symposium on Biomedical Imaging (ISBI)},
year={2023},
organization={IEEE}
}
@inproceedings{gupta2021improved,
title={Improved brain age estimation with slice-based set networks},
author={Gupta, Umang and Lam, Pradeep K and Ver Steeg, Greg and Thompson, Paul M},
booktitle={2021 IEEE 18th International Symposium on Biomedical Imaging (ISBI)},
pages={840--844},
year={2021},
organization={IEEE}
}
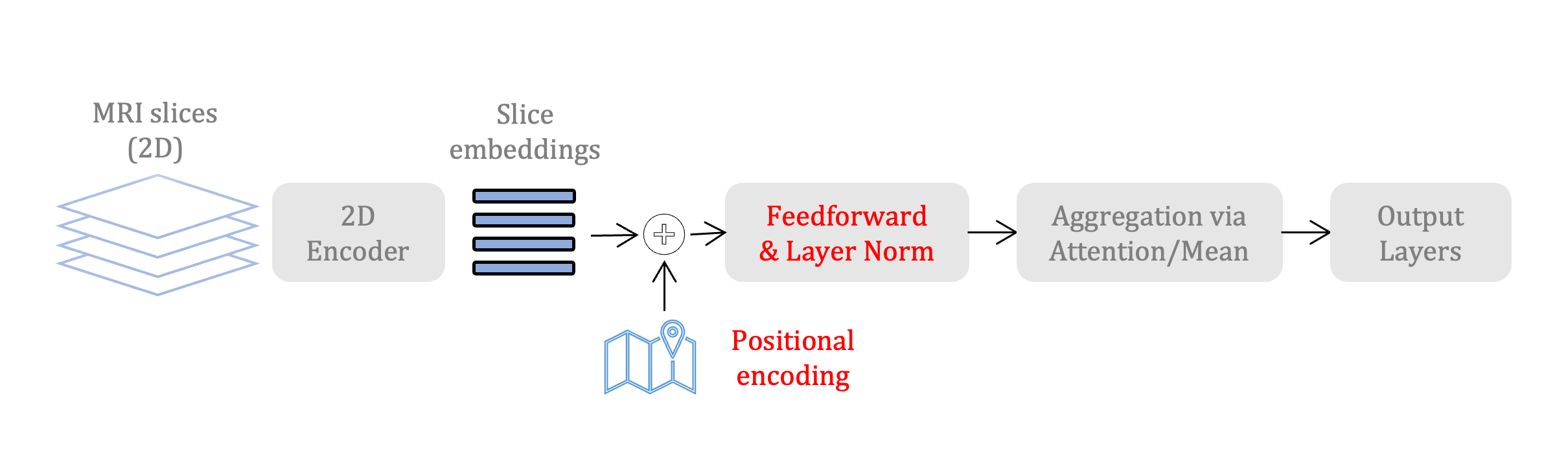 We proposed a new architecture for making predictions from 3D MRIs, which works by encoding each 2D slice in an MRI with a deep 2D-CNN model and combining the information from these 2D-slice encodings by using set networks or permutation invariant layers or operations such as mean. In the ISBI 2021 paper, we performed experiments on brain age prediction using the UK Biobank dataset. We showed that the model with the permutation invariant aggregation layers trains faster and provides better predictions than the other state-of-the-art approaches.
We proposed a new architecture for making predictions from 3D MRIs, which works by encoding each 2D slice in an MRI with a deep 2D-CNN model and combining the information from these 2D-slice encodings by using set networks or permutation invariant layers or operations such as mean. In the ISBI 2021 paper, we performed experiments on brain age prediction using the UK Biobank dataset. We showed that the model with the permutation invariant aggregation layers trains faster and provides better predictions than the other state-of-the-art approaches.
In the ISBI 2023 paper, we introduce positional encodings to preserve spatial information about the ordering of the slices that can be removed due to permutation invariant operations. And we demonstrate the usefulness of the models pretrained on natural images (2D images) by using ImageNet pretrained ResNet models as the slice encoders. We show results on brain age prediction using the UK Biobank dataset and Alzheimer's disease diagnosis on the ADNI dataset.
The code is tested with Python 3.8, but it should work with Python 3.7 or
higher.
See requirements.txt for the package requirements.
We have used UK Biobank & ADNI MRI scans for training. The final dimension of the images is 91×109×91, and they are loaded via nibabel. Our code requires the CSV files of the train/test/valid dataset. See the data folder for how to set up the CSV files. For more details about data and training setup, see our paper.
See the src/shell folder to reproduce the results in the paper.
We have organized the commands according to the tables in the paper.
See config/ and src/scripts/main.py to run change or modify params
for training or evaluation.
Our proposed architecture is in src/arch/ukbb/brain_age_slice_set.py.