Transpharmer: Accelerating Discovery of Novel and Bioactive Ligands With Pharmacophore-Informed Generative Models
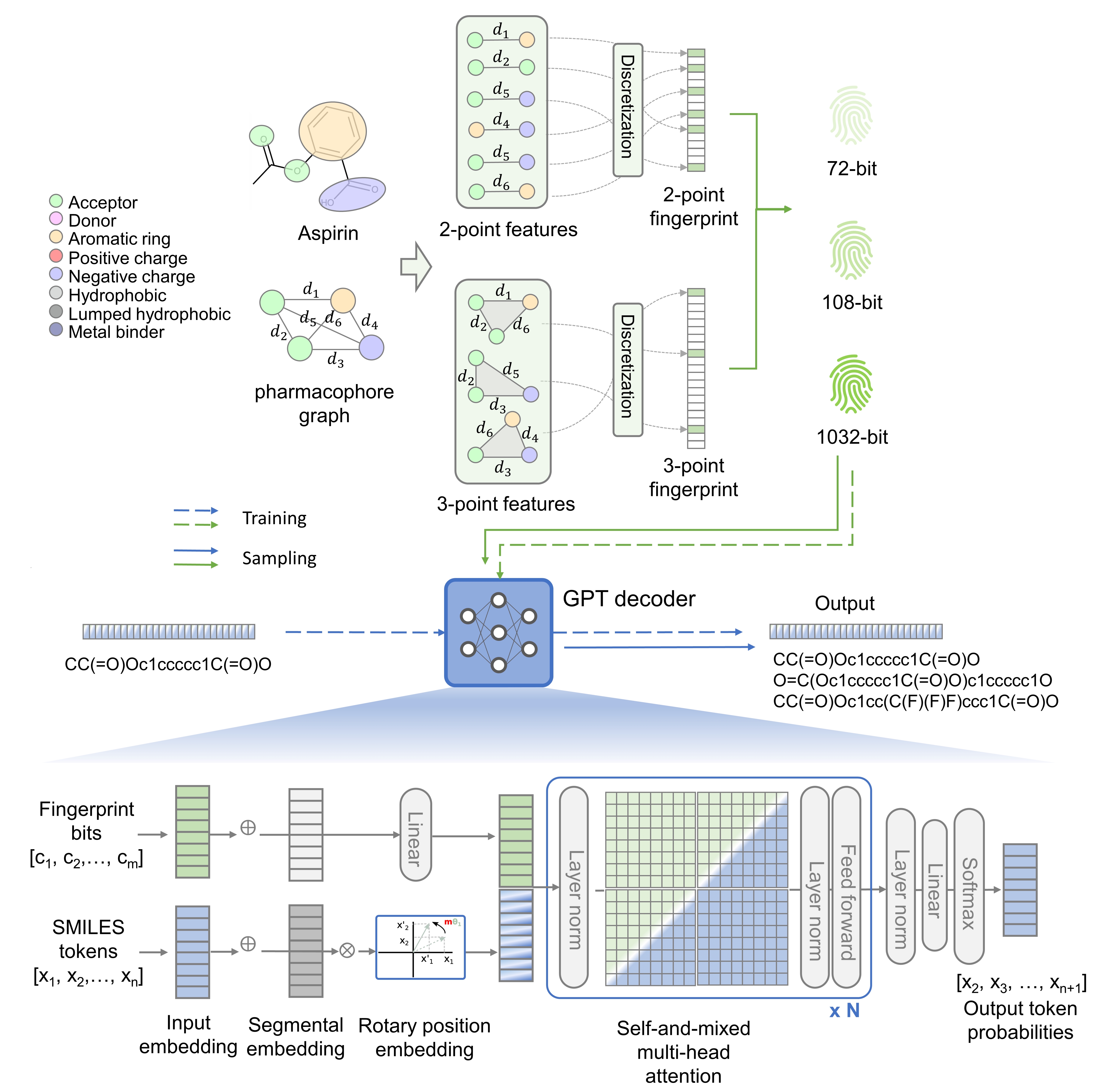
TransPharmer is an innovative generative model that integrates interpretable topological pharmacophore fingerprints with generative pre-training transformer (GPT) for de novo molecule generation. TransPharmer can be used to explore pharmacophorically similar and structurally diverse ligands and has been successfully applied to design novel kinase inhibitors with low-nanomolar potency and high selectivity. The workflow of TransPharmer is illustrated in the figure below.
For more details, please refer to the arXiv paper.
If you find TransPharmer useful, please consider citing us as:
@article{xie2024accelerating,
title={Accelerating Discovery of Novel and Bioactive Ligands With Pharmacophore-Informed Generative Models},
author={Xie, Weixin and Zhang, Jianhang and Xie, Qin and Gong, Chaojun and Xu, Youjun and Lai, Luhua and Pei, Jianfeng},
journal={arXiv preprint arXiv:2401.01059},
year={2024}
}
TransPharmer was tested on the environment with the following installed packages or configurations.
- python 3.9
- torch=1.13.1
- cuda 11.7 (Nvidia GeForce RTX 3090. GPU memory size: 24 GB)
- rdkit=2022.9.3
- scipy=1.8.0
- numpy=1.23.5
- einops==0.6.0
- fvcore==0.1.5.post20221221
- guacamol=0.5.2 (optional)
- tensorflow=2.11.0 (required by and compatible with guacamol 0.5.2)
Here is the step-by-step process to reproduce TransPharmer's working environment:
- Create conda environment and activate:
conda create -n transphamer python=3.9- Install pytorch:
conda install pytorch==1.13.1 pytorch-cuda=11.7 -c pytorch -c nvidia
- Install other requirements using
mamba: first install mamba following the tutorial. (Do not forget to change channels as required.)
mamba env update -n transpharmer --file other_requirements.ymlThe reason to use this hybrid installation process is that conda can be rather annoying and time-consuming to solve environment and settle all requirements. mamba is more faster.
- (Optional) run GuacaMol benchmarking: need to adjust some packages in order to be compatible with GuacaMol. (GuacaMol will automatically install the latest version of TensorFlow, which can be problematic.)
Manual upgrade or downgrade with pip to fulfill:
- tensorflow=2.11.0
- scipy=1.8.0
- numpy=1.23.5
We used the GuacaMol pre-built datasets to train and validate our model, which can be downloaded here. (However, if the links are inaccessible temporarily, we also provide a copy here, since GuacaMol is under MIT license.)
cd transpharmer-repo/
unzip guacamol.zip
ls data/Pretrained TransPharmer model weights can be downloaded here. The organization of the downloaded directory is described as follows:
cd transpharmer-repo/
unzip TransPharmer_weights.zip
ls weights/
weights/
guacamol_pc_72bit.pt: trained with 72-bit pharmacophore fingerprints of GuacaMol compounds;
guacamol_pc_80bit.pt: trained with 72-bit pharmacophore fingerprints concated with feature count vectors;
guacamol_pc_108bit.pt: trained with 108-bit pharmacophore fingerprints;
guacamol_pc_1032bit.pt: trained with 1032-bit pharmacophore fingerprints;
guacamol_nc.pt: unconditional version trained on GuacaMol;
moses_nc.pt: unconditional version trained on MOSES;
We provide several configuration files (*.yaml) for different utilities:
- generate_pc.yaml (configuration of pharmacophore-conditioned generation)
- generate_nc.yaml (configuration of unconditional/no-condition generation)
- benchmark.yaml (configuration of benchmarking)
- train.yaml (configuration of training)
Each yaml file contains model and task-specific parameters. See explanation.yaml and Tutorial for more details.
To train your own model, use the following command line:
python train.py --config configs/train.yaml
To generate molecules similar to input reference compounds in terms of pharmacophore, usually some known actives, use the following command line:
python generate.py --config configs/generate_pc.yaml
Generated SMILESs are saved in the user-specified csv file. The generated csv file has two columns (Template and SMILES) in pharmacophore-conditioned (pc) generation mode (or one column SMILES in unconditional (nc) generation mode).
A demo is provided in the Tutorial.
To benchmark our unconditional model with GuacaMol, run the following command line:
python benchmark.py --config configs/benchmark.yaml
To benchmark our unconditional model with MOSES, generate samples using benchmark.py or generate.py with generate_nc.yaml config and compute all MOSES metrics following their guidelines.
To evaluate get_metrics in benchmark.py with generated csv file.
To reproduce the results of case study of designing PLK1 inhibitors in our paper, run the following command:
python generate.py --config configs/generate_reproduce_plk1.yamlAfter filtering out invalid and duplicate SMILES, one should be able to find exactly the same structure as lig-182 (corresponds to the most potent synthesized compound IIP0943) and structures with identical BM scaffolds to lig-3, lig-524 and lig-886.
- Youjun Xu (xuyj@iipharma.cn)
- Weixin Xie (xiewx@pku.edu.cn)
- Jianhang Zhang (zhangjh@iipharma.cn)
MIT license. See LICENSE for more details.
We thank authors of the following repositories to share their codes: