Co-BioNet: Uncertainty-Guided Dual-Views for Semi-Supervised Volumetric Medical Image Segmentation
This repo contains the supported pytorch code and configuration files to reproduce results of Uncertainty-Guided Dual-Views for Semi-Supervised Volumetric Medical Image Segmentation Article.
Abstract
The tremendous progress in deep learning has resulted in accurate models in the field of medical AI. However, these deep learning models usually require large amounts of annotated data for training, which is prone to human biases and quite often unavailable for dense prediction tasks such as image segmentation. Inspired by semi-supervised deep learning learning algorithms, which make use of both labeled and unlabeled data for training, we propose a dual-view framework based upon adversarial learning for image segmentation. In doing so, we employ two critics to allow each view to learn from high-confidence predictions of the other view by determining the uncertainty of generated predictions using entropy calculation. Further, to jointly learn the dual views and their critics, we formulate the learning problem as a min-max problem. We analyze and contrast our proposed method against state-of-the-art baselines, both qualitatively and quantitatively, on the National Institutes of Health (NIH) pancreas CT dataset and the Left Atrial Segmentation Challenge (LA) MRI dataset and demonstrate that the proposed semi-supervised method substantially outperforms the competing baselines while achieving competitive performance compared to fully-supervised counterparts. We hypothesize that uncertainty guided co-training framework can make two competing neural networks that are more robust to data artefacts and has the ability to generate multiple plausible segmentation masks that can be helpful in semi automated segmentation process.
Link to full paper:
To be Added
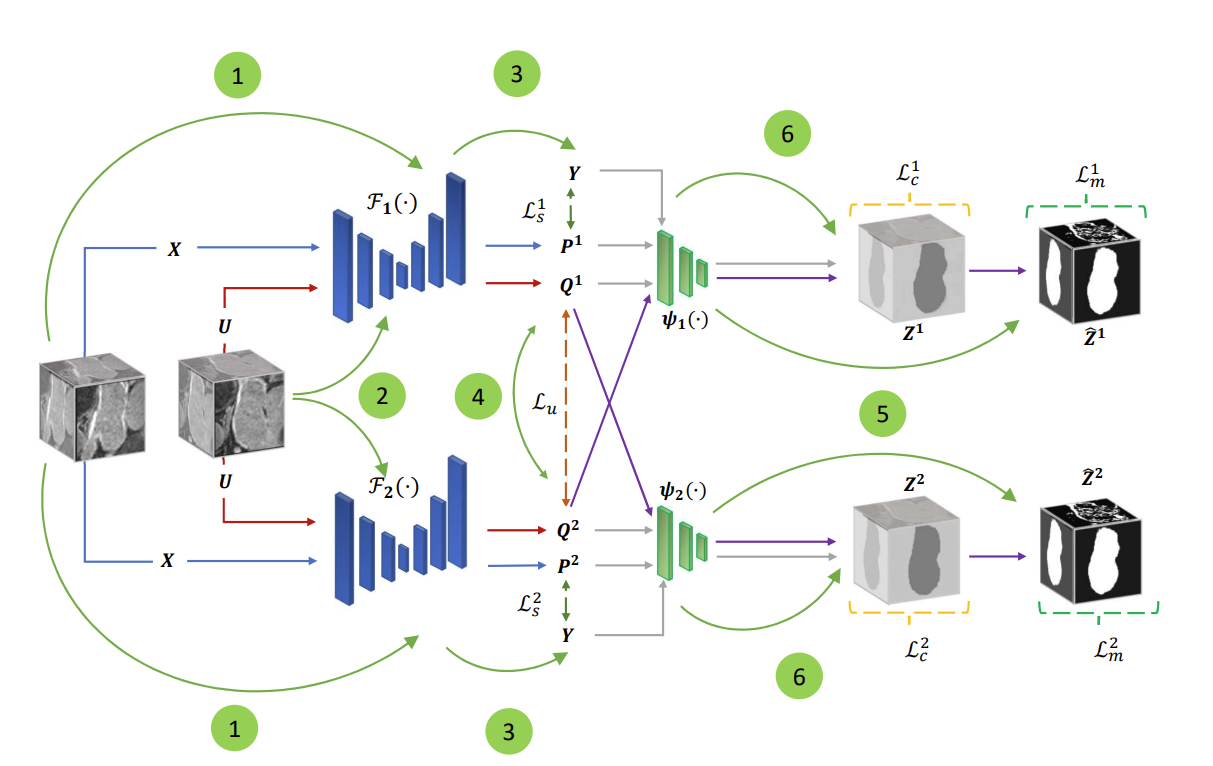
Proposed Architecture
System requirements
Under this section, we provide details on environmental setup and dependencies required to train/test the Co-BioNet model.
This software was originally designed and run on a system running Ubuntu (Compatible with Windows 11 as well).
All the experiments are conducted on Ubuntu 20.04 Focal version with Python 3.8.
To train Co-BioNet with the given settings, the system requires a GPU with at least 24GB. All the experiments are conducted on Nvidia RTX 3090 single GPU.
(Not required any non-standard hardware)
To test model's performance on unseen test data, the system requires a GPU with at least 4 GB.
Create a virtual environment
virtualenv -p /usr/bin/python3.8 venv
source venv/bin/activateInstallation guide
- Install torch :
pip3 install torch==1.10.2+cu113 torchvision==0.11.3+cu113 torchaudio==0.10.2+cu113 -f https://download.pytorch.org/whl/cu113/torch_stable.html- Install other dependencies :
pip install -r requirements.txtTypical Install Time
This depends on the internet connection speed. It would take around 15-30 minutes to create environment and install all the dependencies required.
Dataset Preparation
The experiments are conducted on two publicly available datasets,
- National Institutes of Health (NIH) Panceas CT Dataset : https://wiki.cancerimagingarchive.net/display/Public/Pancreas-CT
- 2018 Left Atrial Segmentation Challenge Dataset : http://atriaseg2018.cardiacatlas.org
Pre-processed data can be found in folder data.
Trained Model Weights
Download trained model weights from this shared drive link, and put it under folder code/model
Running Demo
Demonstration is created on generating segmentation masks on a sample of unseen Pancreas CT with trained torch models on 10% and 20% Labeled Pancreas CT and Left Atrial MRI data. You can run the given python notebook in the demo folder.
Train Model
- To train the model for Pancreas CT dataset on 10% Lableled data
cd code
nohup python train_cobionet_PANCREAS.py --labelnum 6 --lamda 1.0 --consistency 1.0 --mu 0.01 --t_m 0.2 --max_iteration 15000 &> pa_10_perc.out &- To train the model for Pancreas CT dataset on 20% Lableled data
cd code
nohup python train_cobionet_PANCREAS.py --labelnum 12 --lamda 1.0 --consistency 1.0 --mu 0.01 --t_m 0.2 --max_iteration 15000 &> pa_20_perc.out &- To train the model for Left Atrial MRI dataset on 10% Lableled data
cd code
nohup python train_cobionet_LA.py --labelnum 8 --lamda 0.8 --consistency 1.0 --mu 0.01 --t_m 0.3 --max_iteration 15000 &> la_10_perc.out &- To train the model for Left Atrial MRI dataset on 20% Lableled data
cd code
nohup python train_cobionet_LA.py --labelnum 16 --lamda 0.8 --consistency 1.0 --mu 0.01 --t_m 0.3 --max_iteration 15000 &> la_20_perc.out &It would take around 5 hours to complete model training.
Test Model
- To test the model 1 for Pancreas CT dataset on 10% Lableled data
cd code
python eval_3d.py --dataset_name Pancreas_CT --labelnum 6 --model_num 1- To test the ensemble model for Pancreas CT dataset on 10% Lableled data
cd code
python eval_3d_ensemble.py --dataset_name Pancreas_CT --labelnum 6- To test and get best segmentation masks that are more closer to ground truth annotations out of model 1, model 2 and the ensemble model for Pancreas CT dataset on 10% Lableled data
cd code
python eval_get_best.py --dataset_name Pancreas_CT --labelnum 6Acknowledgements
This repository makes liberal use of code from SASSNet, UAMT, DTC and MC-Net