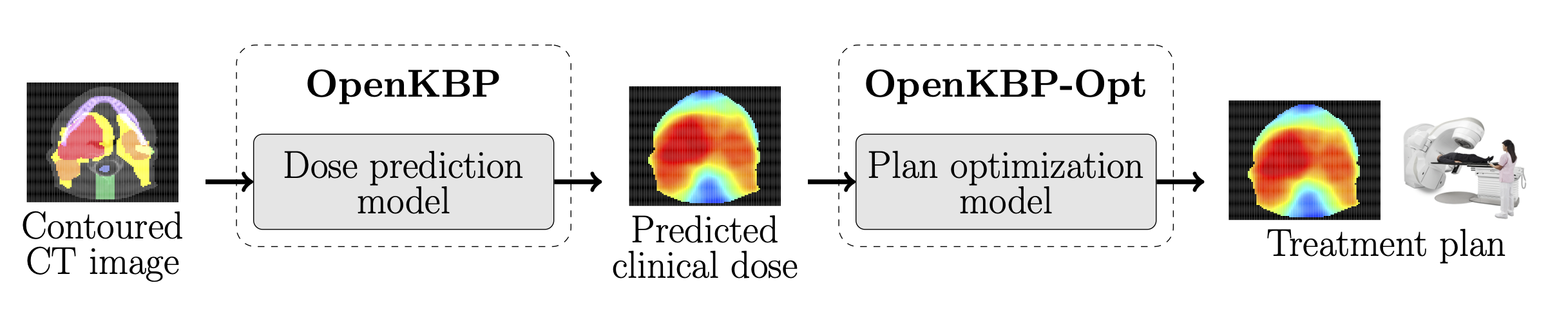
The open-kbp-opt repository is part of the OpenKBP-Opt project, which is an extension of the OpenKBP Grand Challenge. This repository provides code and open data to reproduce the experiments from our paper, however, we encourage others to modify our experiments to extend this work.
Please use our paper as the citation for this dataset or code repository:
A. Babier, R. Mahmood, B. Zhang, V.G.L. Alves, A.M. Barragán-Montero, J. Beaudry, C.E. Cardenas, Y. Chang, Z. Chen, J. Chun, K. Diaz, H.D. Eraso, E. Faustmann, S. Gaj, S. Gay, M. Gronberg, B. Guo, J. He, G. Heilemann, S. Hira, Y. Huang, F. Ji, D. Jiang, J.C.J. Giraldo, H. Lee, J. Lian, S. Liu, K. Liu, J. Marrugo, K. Miki, K. Nakamura, T. Netherton, D. Nguyen, H. Nourzadeh, A.F.I. Osman, Z. Peng, J.D.Q. Muñoz, C. Ramsl, D.J. Rhee, J.D. Rodriguez, H. Shan, J.V. Siebers, M.H. Soomro, K. Sun, A.U. Hoyos, C. Valderrama, R. Verbeek, E. Wang, S. Willems, Q. Wu, X. Xu, S. Yang, L. Yuan, S. Zhu, L. Zimmermann, K.L. Moore, T.G. Purdie, A.L. McNiven, T.C.Y. Chan, “OpenKBP-Opt: An international and reproducible evaluation of 76 knowledge-based planning pipelines,” Physics in Medicine and Biology, Vol. 67, pp. 185012, 2022.
- Data
- What this code does
- Prerequisites
- Created folder structure
- Getting started
- Running the code
- Citation
The details of the provided data are available in our paper OpenKBP-Opt: An international and reproducible evaluation of 76 knowledge-based planning pipelines. In short, we provide data for 100 patients who were treated for head-and-neck cancer with intensity modulated radiation therapy (IMRT), and they are the same patients as those in the testing set from the OpenKBP Grand Challenge. Every patient in this dataset has a reference plan dose distribution (i.e., a ground truth), 21 predicted dose distributions, a dose influence matrix, CT images, structure masks, a feasible dose mask (i.e., mask of where dose can be non-zero), and voxel dimensions. Each predicted dose distribution was produced by a model that was trained on an out-of-sample training set (n=200 patients) and validation set (n=40 patients) by practitioners who only had the ground truth data (i.e., dose distribution) for the training set. Please note that researchers who are interested in developing their own dose prediction methods should use the open-kbp repository.
This code will solve optimization models to generate radiotherapy treatment plans. This repository includes two scripts and eight other PY files that contain functions and classes that are specific to this repository.
These two main scripts will complete all experiments and produce all visualizations/tables from our paper.
- main.py: Solves an optimization model on all patients (n=100) in the dataset and each of their high-quality predictions (n=19), which is a total of 7600 optimization models. On most computers this will take at least 1 month to complete (outputs are stored as part of the complementary dataset), however, the solutions are available in the dataset associated with this repository.
- main_analysis.py: Generates the tables and plots from the complementary paper.
We summarize the functionality of the eight files that contain our repository specific functions and classes. More details are provided in the files/functions themselves.
- analysis_data_prep.py: Contains several functions that prepare data for easier analysis
- constants_class.py: Contains the ModelParameters class, which has several attributes that are used in many of the functions in this repository.
- data_loader.py: Contains the DataLoader class, which loads the data from the dataset in a standard format.
- dose_evaluation_class.py: Contains the EvaluateDose class, which is used to evaluate the competition metrics.
- general_functions.py: Contain several functions with a variety of purposes.
- optimizer.py: Contains the PlanningModel class, which constructs and solves each optimization model.
- plotting.py: Contains functions to construct several of the plots in the complementary paper.
- resources.py: Contains the Patient class, which is used to store and manipulate patient data (e.g., generating optimization structures)
The following are required to run the given scripts.
- Linux
- Python 3.7.5
- Gurobi 9.1.2 installed with license
Gurobi can be installed by following the first 5 pages of these instructions. Academics can use Gurobi for free by downloading the solver and requesting an Academic License.
This code was only tested with Gurobi, however, we construct the models via Google OR-Tools. As a result, this code should also work with other solvers that are compatible with Google OR-Tools (e.g., CPLEX, SCIP, GLPK, GLOP) by changing the optimization solver on Line 53 in optimizer.py.
This repository will use a file structure that branches from a directory called open-kbp-opt-data. All the reference patient data (e.g., reference dose, contours) are stored in reference-plans. The predictions that were generated during the OpenKBP Grand Challenge are stored in paper-predictions, and the plans that we generated using those predictions are stored in paper-plans. The paper-plans directory also has a subdirectory for each optimization model (identified by the name of the appropriate optimization model). Lastly, results-data contains the summary statistics from the reference plans/predictions/KBP generated plans that are used to generate the aggregate results (e.g., plots, tables in paper), which are stored in results.
open-kbp-opt-data
├── reference-plans
│ ├── pt_*
│ │ ├── *.csv
├── paper-predictions
│ ├── set_*
│ │ ├── pt_*.csv
├── paper-plans
│ ├── model_names
│ │ ├── plan-dose
│ │ │ ├── pt_*.csv
│ │ ├── plan-fluence
│ │ │ ├── pt_*.csv
│ │ ├── plan-gap
│ │ │ ├── pt_*.csv
│ │ ├── plan-weights
│ │ │ ├── pt_*.csv
├── results-data
│ ├── *
└── results
├── *
-
Make a virtual environment and activate it
virtualenv -p python3 open-kbp-opt-venv source open-kbp-opt-venv/bin/activate -
Clone this repository, navigate to its directory, and install the requirements.
git clone https://github.com/ababier/open-kbp-opt cd open-kbp-opt pip3 install -r requirements.txt -
Download the data for reference-plans and paper-predictions directories (10.19 GB) from our OneDrive or via the command below
sh data_download_commands/patients_and_predictions.txt -
Optional: Download the data for paper-plans, results-data, and results directories (13.08 GB) from our OneDrive or via the command below
sh data_download_commands/kbp_plans_and_results.txt
Running the code on the intended platform should be straightforward. Any errors are likely the result of data being in an unexpected directory or issues related to Gurobi installation. and running main_analysis.py will generate new results.
Run the main files in your newly created virtual environment.
python3 main.py
python3 main_analysis.py
If the code is running correctly then running main.py should solve an optimization model for each pair of patients and predictions. No optimization will be performed if solutions (i.e., plans) for the optimization model are already saved. For example, if you download the optional data, which contains the solutions to all optimization models solved in the paper) the script will bypass all optimization models because solutions already exist. Note that running the optimization models for all pairs of patients and prediction will take several weeks.
Once solutions exist main_analysis.py can be run to generate summary statistics and plots to summarize the performance of the models. Note that the first run of main analysis will take about an 15 minutes per optimization model to complete because it needs to load and summarize the data for 1900 treatment plans for each optimization model. Following the first run, the summary statistics are saved such that successive runs will take a few seconds.