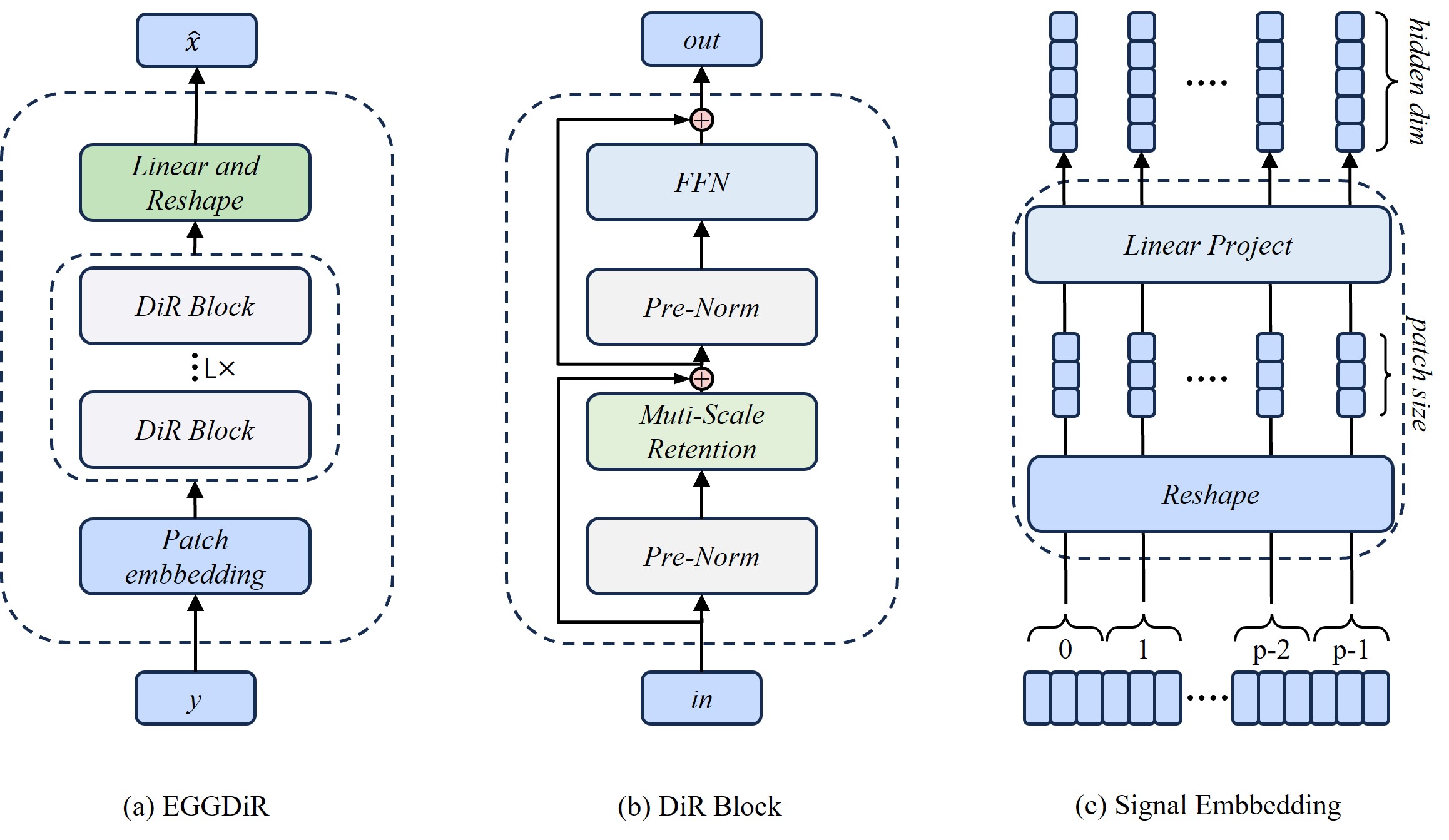
EEGDiR: Electroencephalogram denoising network for temporalinformation storage and global modeling through Retentive Network
Electroencephalogram (EEG) signals are essential in clinical medicine, brain research, and neurological disease studies. However, various physiological and environmental artifacts introduce noise, hindering accurate analysis of brain activity. Recent advancements in deep learning offer promising solutions for enhancing EEG signal denoising compared to traditional methods. In this study, we introduce the Retentive Network architecture from large language model (LLM) to EEG signal denoising, leveraging its robust feature extraction and global modeling capabilities. To adapt Retnet to the one-dimensional nature of EEG signals, we propose a signal embedding method, transforming EEG signals into two dimensions for network input. This integration presents a novel approach to EEG denoising, opening avenues for a profound understanding of brain activities and accurate diagnosis of neurological diseases. In addition, since the creation of deep learning datasets is very time-consuming and cumbersome, we also provide a standardized ready-to-use dataset that has been preprocessed to accelerate the progress of deep learning methods. Based on the dataset we produced and the proposed method, t he experimental results show that the denoising effect is significantly improved compared to the existing methods.
Congratulations! This paper was accepted by Elsevier CIBM.
https://www.sciencedirect.com/science/article/abs/pii/S001048252400711X
dataset is available at: https://huggingface.co/datasets/woldier/eeg_denoise_dataset
The method chosen in this article is to download and extract *.tar.gz to the {your_path}/data directory.
The structure is as follows:
data/dataset/
├── EMG
├── train
├── test
├── EOG
├── train
├── test
├── SS-EOG
├── train
├── test
└── test
The project is built with PyTorch 1.13.1, Python3.10, CUDA11.7. For package dependencies, you can install them by:
pip install -r requirements.txtWe use yaml format configuration files, and import the configuration files via pyyaml.
The configuration files are stored in the {your_path}/config/ directory.
You need to set the path to config in train.py.
The configuration of config.yml is divided into four main sections, Model Configuration, Training Parameters Configuration, Dataset Configuration, Logging Configuration.
#==========================model config================================
model:
# The model initialization parameters are configured
# and passed to the model initialization function as **kwargs.
config:
layers: 1key
hidden_dim: 512
ffn_size: 1024
heads: 8
double_v_dim: False
seq_len: 512
mini_seq: 32
drop_out: 0.1
# Configure the path to the model's package file
#Similar to `from model.retnet.retnet import DiR`
path: model.retnet.retnet
class_name: DiR
# Weight file path, if there is a weight file it will be loaded.
weight_path:
#==========================training config================================
train:
epochs: 5000
batch_size: 1000 # Number of samples in a batch
save_img_rate: 1 # How many rounds to save a step
learning_rate: 0.0005
#==========================dataset config================================
dataset:
train:
dataset_path: ./data/dataset/EMG/train # the path to load train data
test:
dataset_path: ./data/dataset/EMG/test # the path to load test data
#==========================logs=======================================
logs:
name: DiR_4_EOG_pathch16_mini_seq32_hidden_dim512_layer_1_EMGFor the log file, it will be saved in the same directory as train.py by default.
In addition, an extra configuration file is saved. This is used to save the training details.
The structure of the log folder is as follows:
results/DiR_4_EOG_pathch16_mini_seq32_hidden_dim512_layer_1_EMG/
├── img
├── train
├── test
├── logs
├── log.txt
├── weight
├── best.pth
├── Epoch0.pth
└── config.yml
- Single GPU pycharm run
The easiest way is to run
trian.pydirectly under pycham
This library uses the Accelerate library, so you don't need to make any modifications to this training framework. Accelerate will automatically convert your data to the GPU (if supported). 2. Multi-GPU run in CMD
If you want to train on multiple GPUs, you can refer to the following code
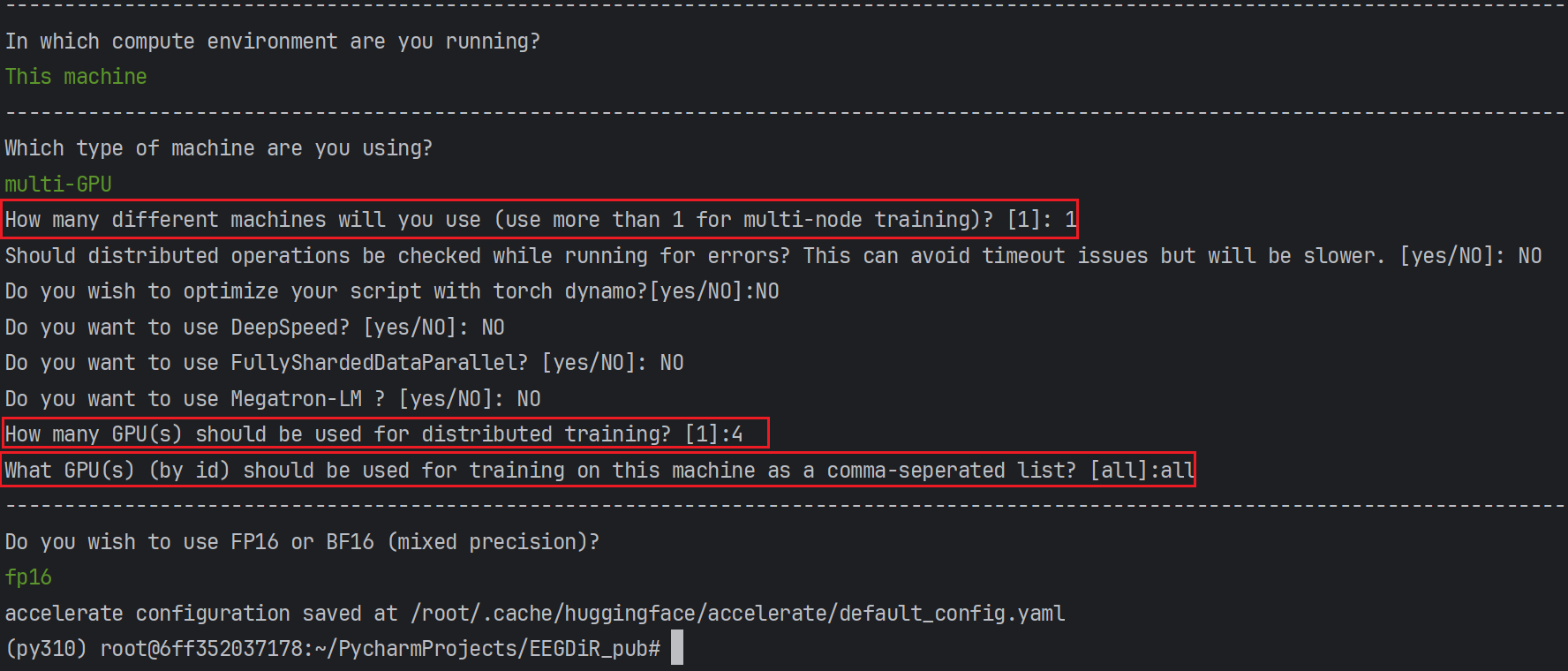
Before running it for the first time, you need to generate a configuration file through accelerate, telling accelerate which GPUs are currently involved in the training.
accelerate configOnce you have configured the accelerate config file, you can view the configuration via accelerate env(Optional).
Multi-GPU training can then be performed by calling train.py.
accelerate launch --gpu_ids=all {your_path}/train.pyFor more details, please refer to the Accelerate website: https://huggingface.co/docs/accelerate/basic_tutorials/launch 3. Multi-GPU run in pycharm
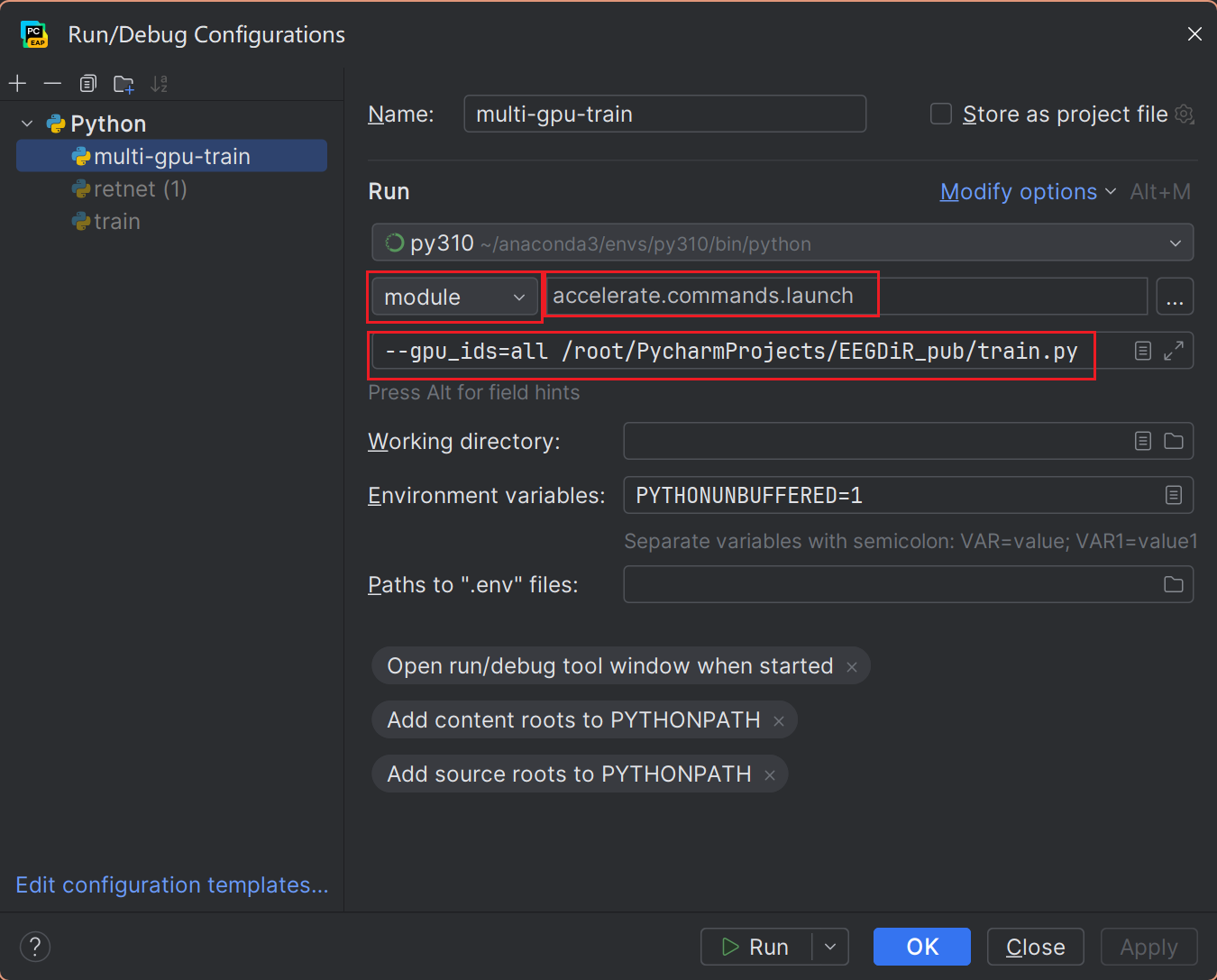
If you find the cmd call scripts inelegant, you can also configure pycharm.
- Create a New Run/Debug Configuration
- Setting the ENV of RUN
- Set the mode to module, and give the model name:
accelerate.commands.launch. - Set the parameter
--gpu_ids=allto set the available GPUs, and specify the script path{your_path}/trian.py. - Note that it is recommended to inform Accelerate of the current device hardware via
accelerate configbefore running.
enjoy!
This training framework is not perfect, I will continue to update it.
If you have any ideas on how to improve this framework please issue or PR.