scDIOR: Single cell RNA-seq Data IO softwaRe
- scDIOR
scDIOR software was developed for single-cell data transformation between platforms of R and Python based on Hierarchical Data Format Version 5 (HDF5). There is a data IO ecosystem composed of two modules, dior and diopy, between three R packages (Seurat, SingleCellExperiment, Monocle) and a Python package (Scanpy).
scDIOR accommodates a variety of data types across programming languages and platforms in an ultrafast way, including single-cell RNA-seq and spatial resolved transcriptomics data, using only a few codes in IDE or command line interface.
Users install and operate scDIOR following two ways:
- The environment is created by
conda createin which scDIOR is installed. - Docker images are available on the jiekailab/scdior-image.
The environment is created by conda create in which dior and diopy are installed.
conda create -n conda_env python=3.8 R=4.0- R installation:
# for R
install.packages('devtools')
devtools::install_github('JiekaiLab/dior')
# or devtools::install_github('JiekaiLab/dior@HEAD')- Python installation:
# for python
pip install diopyIt is recommend to perform scDIOR in docker image, which ensures that the operating environment remains stable. scDIOR image is available on the jiekailab/scdior-image.
Brief description
- We first built the basic jupyter image which based on jupyter/base-notebook (jupyter managing Python and R) and fixuid (fixing user/group mapping issues in containers). This basic image is on jiekailab/scdior-image:base-jupyter-notebook1.0.
- Based on our customized basic image, we built scDIOR image again by
Dockerfile. For the content ofDockerfile, it is at this link.
The current latest image contains the following main analysis platforms and software:
| R | version | Python | version |
|---|---|---|---|
| R | 4.0.5 | Python | 3.8.8 |
| Seurat | 4.0.2 | Scanpy | 1.8.1 |
| SingleCellExperiment | 1.12.0 | scvelo | 0.2.3 |
| monocle3 | 1.0.0 | anndata | 0.7.6 |
| dior | 0.1.5 | diopy | 0.5.2 |
At present, scDIOR is widely compatible with Seurat (v3~v4) and Scanpy (1.4~1.8) in different docker image. We configured multiple version docker image (https://hub.docker.com/repository/docker/jiekailab/scdior-image) to confirm that scDIOR can work well between multiple versions of Scanpy and Seurat.
| Platform | Software | Version | data IO |
|---|---|---|---|
| R | Seurat | v3~v4 | ☑️ |
| Python | Scanpy | v1.4~v1.8 | ☑️ |
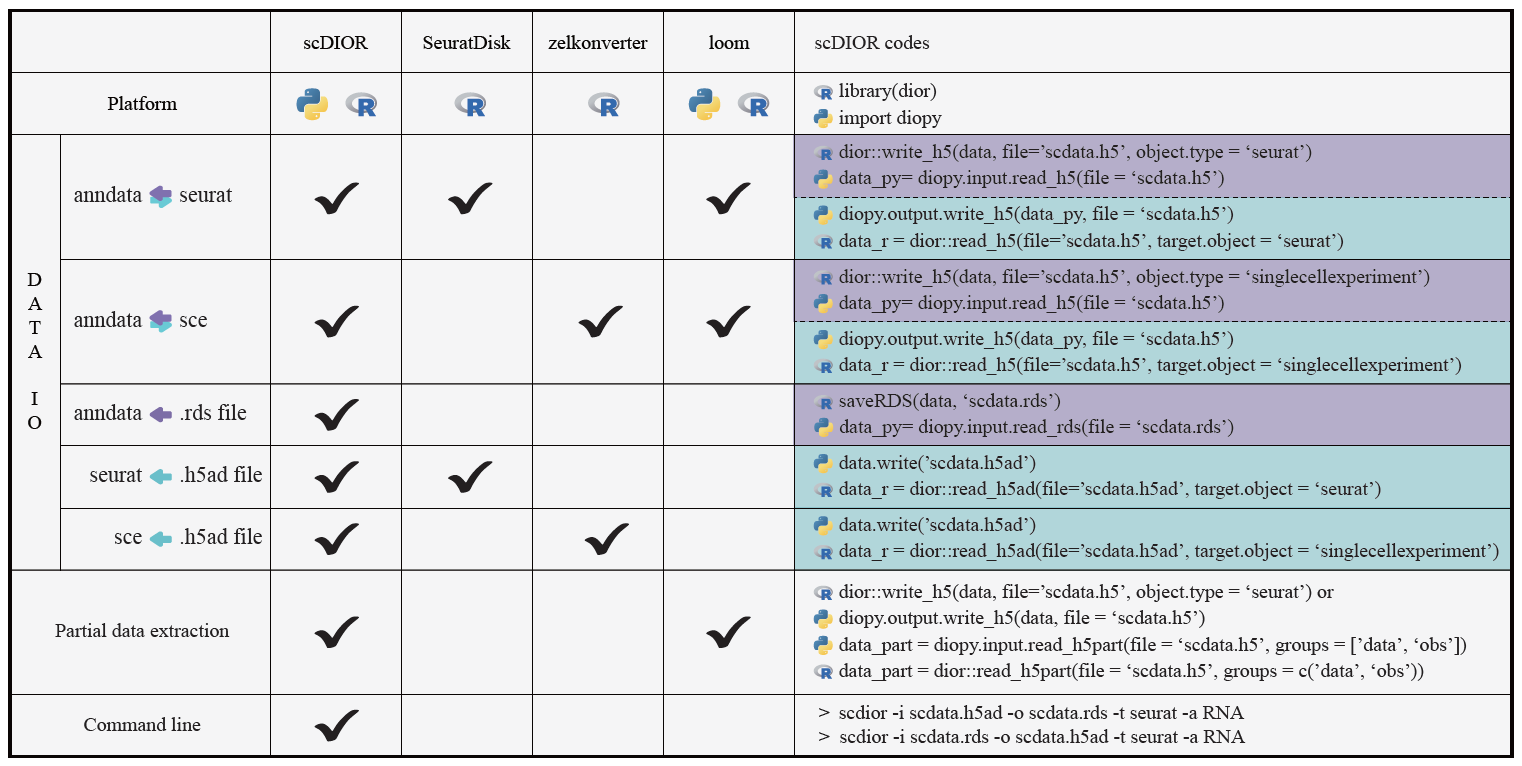
Here, we list several demos to show the powerful performance of scDIOR.
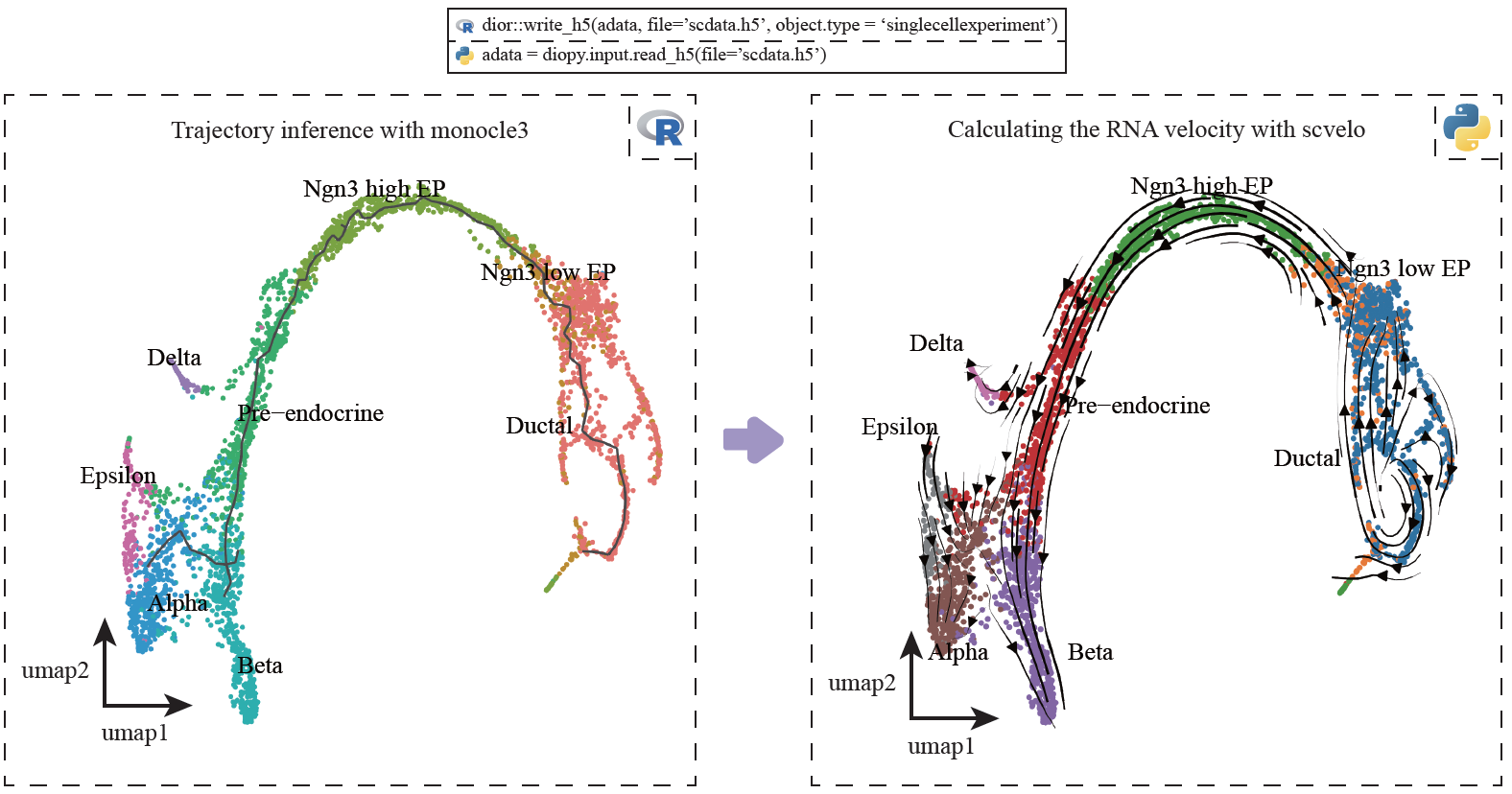
Users can perform trajectory analysis using Monocle3 in R, then transform the single-cell data to Scanpy in Python using scDIOR, such as expression profiles of spliced and unspliced, as well as cell layout. The expression profile can be used to run dynamical RNA velocity analysis and results can be projected on the layout of Monocle3.
Code
# in R
dior::write_h5(data, file='scdata.h5' object.type = 'singlecellexperiment')# in Python
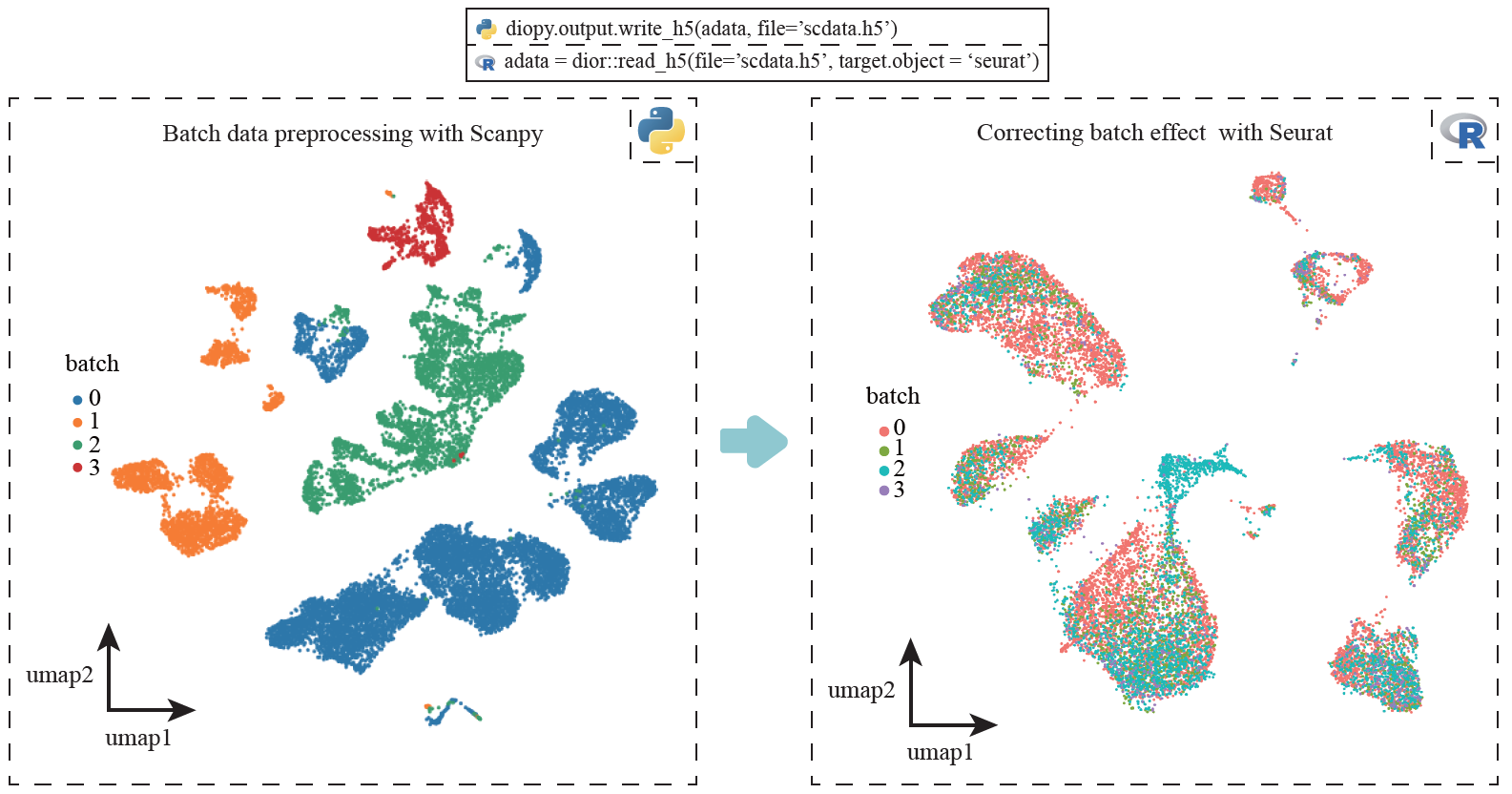
adata = diopy.input.read_h5(file = 'scdata.h5')Users can employ single-cell data processes and normalization method provided by Scanpy, and utilize batches correction method provided by Seurat.
Code
# in python
diopy.output.write_h5(data_py, file = 'scdata.h5')# in R
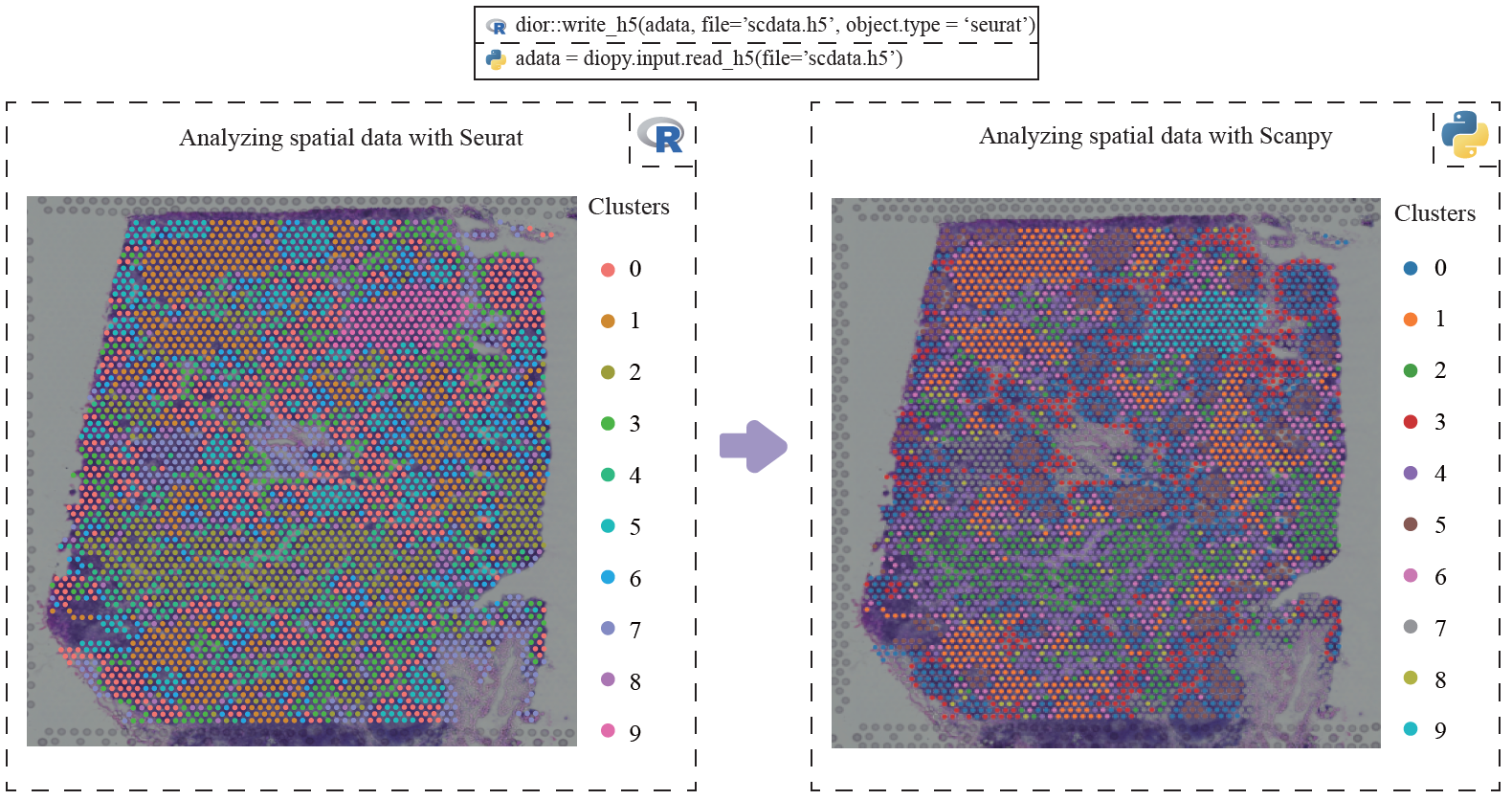
adata = dior::read_h5(file='scdata.h5', target.object = 'seurat')scDIOR supports spatial omics data IO between R and Python platforms.
Code
# in R
dior::write_h5(data, file='scdata.h5', object.type = 'singlecellexperiment')# in Python
adata = diopy.input.read_h5(file = 'scdata.h5')-
The function to load ‘.rds’ file in Python directly;
Code
# in python adata = diopy.input.read_rds(file = './adata_R.rds', object_type='seurat', assay_name='RNA')
-
The function to load ‘.h5ad’ file in R directly;
Code
# in R adata_seurat = read_h5ad(file = './adata_Python.h5ad', target.object = 'seurat', assay_name = 'RNA')
-
Command line
Description
ScDIOR uses the command line to convert different data by calling
scdior.usage: scdior [-h] -i INPUT -o OUTPUT -t TARGET -a ASSAY_NAME-i,--inputThe existing filename for different platforms, such as rds (R) or h5ad (Python).-o,--outputThe filename that needs to be converted, such as from rds to h5ad or from h5ad to rds.-t,--targetThe target object for R, such as seruat or singlecellexperiment.-a,--assay_nameThe primary data types, such as scRNA data or spatial data.Code
$ scdior -i ./adata_test.h5ad -o ./adata_test.rds -t seurat -a RNA
- Our article: https://doi.org/10.1186/s12859-021-04528-3
- jupyter docker stacks:
- fixuid: https://github.com/boxboat/fixuid
- Seurat: https://satijalab.org/seurat/index.html
- monocle3: https://cole-trapnell-lab.github.io/monocle3/
- Scanpy: https://scanpy.readthedocs.io/en/stable/index.html
- Scvelo: https://scanpy.readthedocs.io/en/stable/index.html