--coding by Simon Zhongyuan Tian
• Demo ChIA-view online: https://chia-view.shinyapps.io/ChIA-view-master/.
----a novel visualization pipeline for multiplex chromatin interactions
Simon Zhongyuan Tian, Daniel Capurso, Minji Kim, Byoungkoo Lee, Meizhen Zheng, Yijun Ruan
Recently, we developed ChIA-Drop1, a novel experimental method for detecting multiplex chromatin interactions with single-molecule precision via droplet-based and barcode-linked sequencing. ChIA-DropBox2 a novel toolkit for analyzing and visulizing multiplex chromatin interactions, which includes: a ChIA-DropBox data processing pipeline3 and a visulizing tool ChIA-View4.
1 Meizhen Zheng, Simon Zhongyuan Tian, Daniel Capurso, Minji Kim, Rahul Maurya, Byoungkoo Lee, Emaly Piecuch et al. "Multiplex chromatin interactions with single-molecule precision." Nature 566, 558 (2019). & GSE109355
3 https://github.com/TheJacksonLaboratory/ChIA-DropBox.git
4 https://github.com/TheJacksonLaboratory/ChIA-view.git
- R (v3.5.2)
- RStudio (v1.1.456 )
ChIA-View needs many R packages, which will be automaticlly installed at the first run:
bindrcpp (v0.2.2), gggenes (v0.3.2), data.table (v1.11.8), plotly (v4.8.0), xtable (v1.8-3), rgdal (v1.3-6), sp (v1.3-1), leaflet (v2.0.2), shinyWidgets (v0.4.4), shinyjs (v1.0.1.9004), shinydashboard (v0.7.1), colourpicker (v1.0.2), tidyr (v0.8.2), stringr (v1.3.1), sitools (v1.4), cowplot (v0.9.4), ggrepel (v0.8.0), ggforce (v0.1.3), bedr (v1.0.4), pheatmap (v1.0.12), dplyr (v0.7.8), gridExtra (v2.3), reshape2 (v1.4.3), ggplot2 (v3.1.0), RColorBrewer (v1.1-2), shiny (v1.2.0)
By typing following code in RStudio on your computer, you can easily run ChIA-view.
if (!require("shiny")) install.packages("shiny")
runGitHub("ChIA-view_v1.0", "TheJacksonLaboratory")
Or you can download ChIA-View to your local machine from github and run in RStudio.
git clone https://github.com/TheJacksonLaboratory/ChIA-view.git
When we first time run this server, it may cause a little bit longer time to install necessary packages.
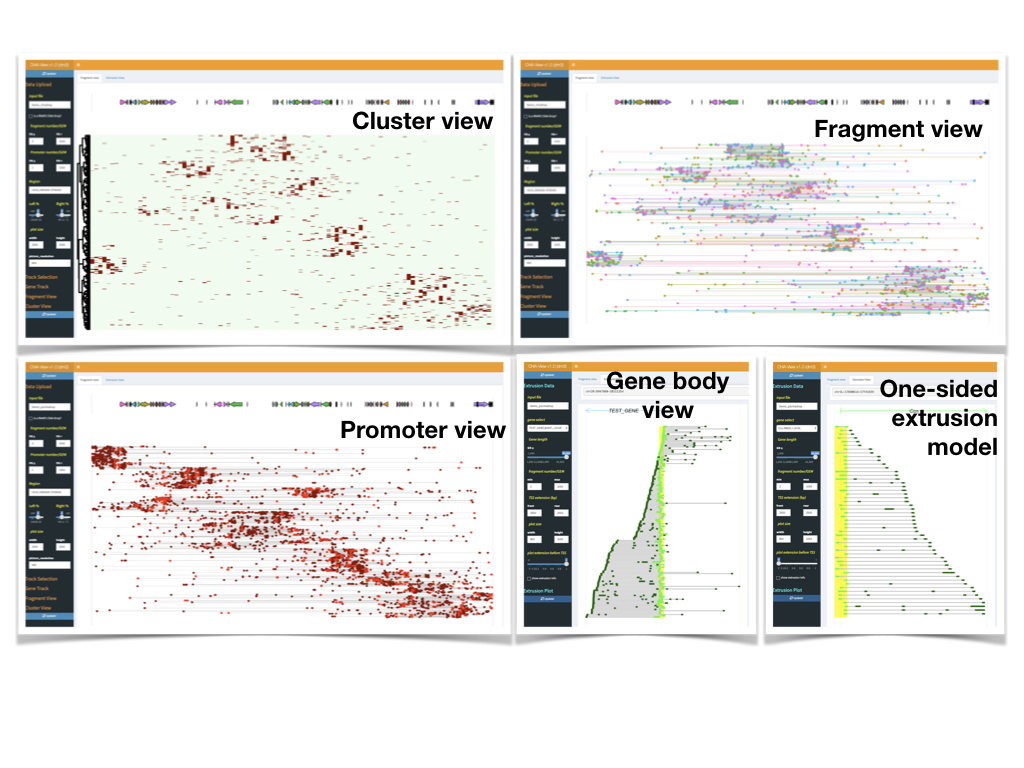
- For ChAI-Drop data, only Cluster VIew and Fragment View are avaiable.
- For RNAPII ChIA-Drop data, Cluster VIew, Fragment View, PE VIew and Extrusion View are all aviable.
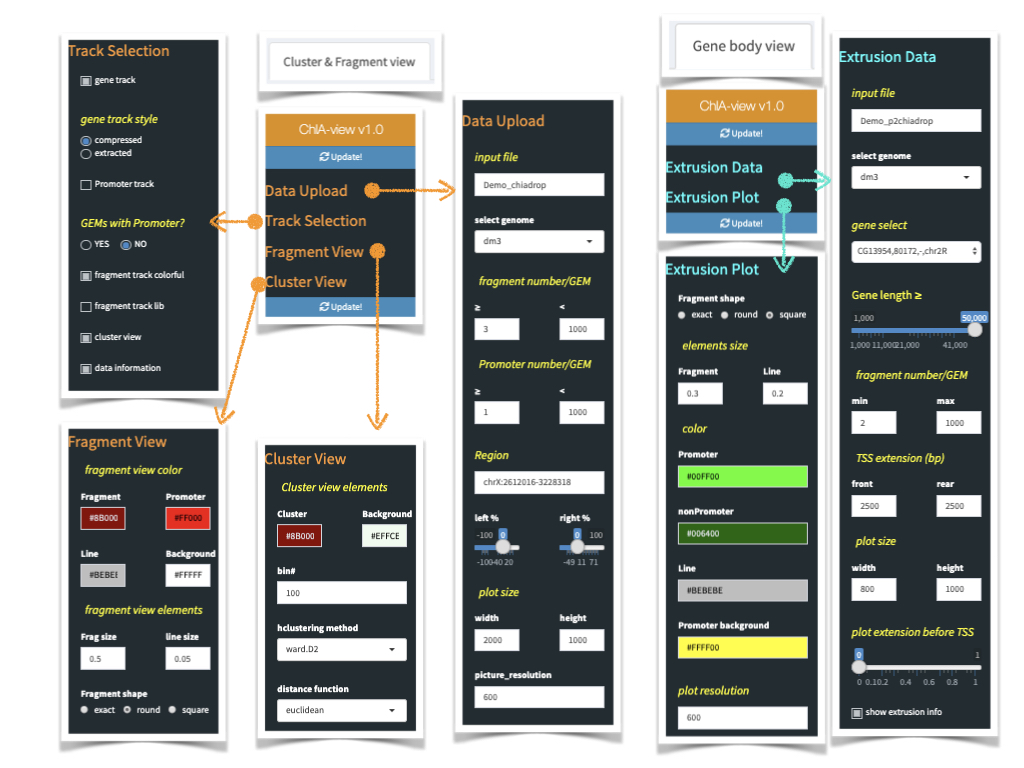
1. Click Update button each time, after you adjusted all/any parameters.
2. Type in folder name of the data you want to upload in "Data Upload/input file" widget.
-
e.g. ChIA-Drop Demo data: type in "Demo_chiadrop"
-
e.g. RNAP2 ChIA-Drop Demo data: type in "Demo_p2chiadrop"
* .region --- Input file for ChIA-Drop data visulized in Cluster view and Fragment view,in which one line represents one fragment; fragments are of a same complex owning same GEMID. One of the standard output of ChIA-DropBox data processing pipeline.
| Chrom | Start | End | Frag_num/GEM | GEM_ID |
|---|---|---|---|---|
| chrX | 16274973 | 16275587 | 2 | SHG0055H-1000-10000046-AAACACCAGTAACGATBX1-M02838-FA-1-0 |
* .region.PEanno --- Input file for RNAP2-ChIA-Drop data visulized in Promoter view and Gene body view, with annotation of fragments by promoter or non-promoter. One of the standard output of ChIA-DropBox data processing pipeline.
| Chrom | Start | End | Frag_num/GEM | GEM_ID | PEanno |
|---|---|---|---|---|---|
| chrX | 16274973 | 16275587 | 2 | SHG0055H-1000-10000046-AAACACCAGTAACGATBX1-M02838-FA-1-0 | P |
| chrX | 16285880 | 16286464 | 2 | SHG0055H-1000-10000046-AAACACCAGTAACGATBX1-M02838-FA-1-0 | E |
We supply a tool (create_input.R) to divide .region or .region.PEanno file from ChIA-DrpBox data processing pipeline to chromosomes specific sub-sets for ChIA-view inputing.
- Copy .region or .region.PEanno file to ChIA-view folder
- Open create_input.R in RStudio
- Modify input file name and output folder name.
IN="Sample.region"
OUT="Sample_SUBRGN"- Then run it
- After a while you could find a folder named Sample_SUBRGN, and using this name for ChIA-view input.
Sample_SUBRGN/
- chr2L.SUBRDS
- chr2R.SUBRDS
- chr3L.SUBRDS
- chr3R.SUBRDS
- chr4.SUBRDS
- chrX.SUBRDS