paleoMap is a package that combines paleomaps from GPlates with fossil records from Paleobiology Database. It can be used to download shapefiles with reconstructions of the past configuration of the continents, and to generate paleodiversity maps.
Install
Install paleoMap from CRAN
install.packages("paleoMap")
library(paleoMap)Install paleoMap developing version from github
install.packages("devtools")
library(devtools)
install_github("macroecology/paleoMap")
library(paleoMap)General overview
paleoMap version 0.1 has 3 functions for getting and visualising paleogeographical maps and fossil data, 4 functions for constructing diversity rasters (paleorichness and Shannon paleodiversity) and 2 functions for creating diversity matrix (based on localities or cells) and 2 functions for getting latitudinal paleodiversity gradients.
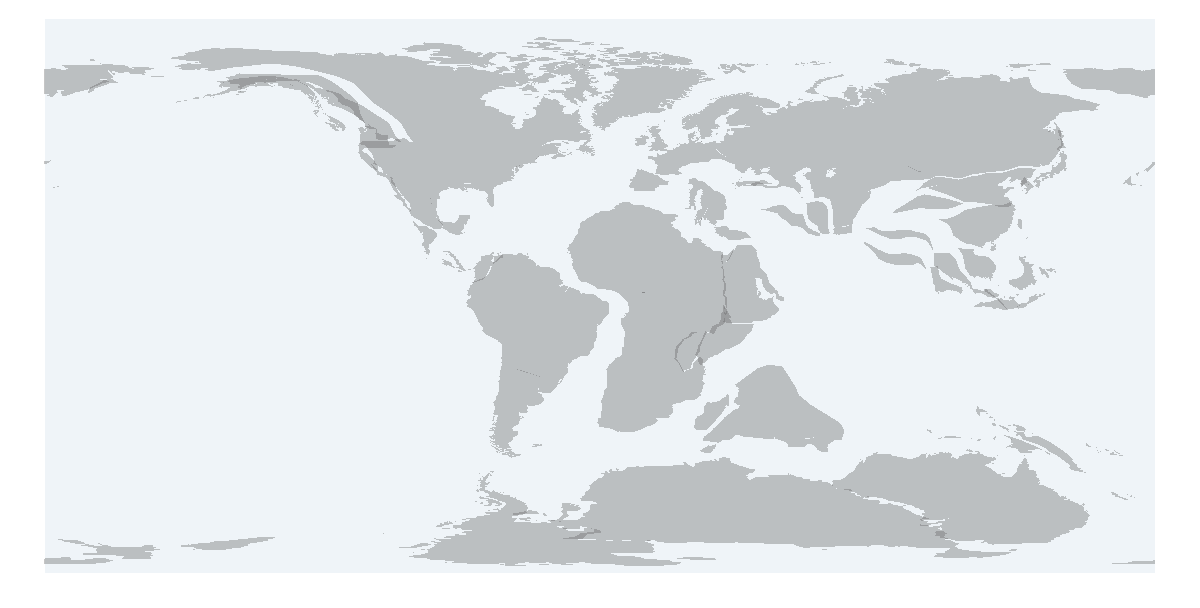
pm_getmap returns the shapefile of a choosen paleogeographical time interval and a plot
> shape <- pm_getmap(interval="Cretaceous")
> shapeclass : SpatialPolygonsDataFrame
features : 86
extent : -180, 180, -88.8737, 83.6951 (xmin, xmax, ymin, ymax)
coord. ref. : NA
variables : 13
names : PLATEID1, GPGIM_TYPE, TYPE, FROMAGE, TOAGE, NAME, DESCR, FEATURE_ID, PLATEID2, L_PLATE, R_PLATE, RECON_METH, SPREAD_ASY
min values : 0, gpml:UnclassifiedFeature, NA, 0, 0, NA, NA, GPlates-052973e4-ed8e-43bf-85b4-aa178e2242a8, 0, 0, 0, NA, 0
max values : 0, gpml:UnclassifiedFeature, NA, 0, 0, NA, NA, GPlates-ffbfeec0-bf61-4769-ac12-bf43ec206a14, 0, 0, 0, NA, 0 pm_getdata returns a dataframe with the selected fossil occurrences, downloaded from the Paleobiology Database
> data <- pm_getdata (base_name="Testudines", interval="Paleocene")
> head(data)
occurrence_no matched_name matched_rank matched_no early_interval late_interval paleolng paleolat geoplate
40165 Peritresius ornatus species 173397 Thanetian <NA> -44.51 40.13 109
40166 Rhetechelys platyops species 128351 Thanetian <NA> -44.51 40.13 109
149344 Aspideretoides virginianus species 316507 Thanetian <NA> -47.60 38.76 109
205060 Judithemys backmani species 253836 Paleocene <NA> -67.18 57.12 101
205061 Trionychinae subfamily 58166 Paleocene <NA> -67.18 57.12 101
281674 Taphrosphys sulcatus species 131500 Late Maastrichtian Early Paleocene -39.82 40.92 109
genus family order class phylum genus_no family_no order_no class_no phylum_no
Peritresius Pancheloniidae Testudines Reptilia Chordata 36360 165650 56475 36322 33815
Rhetechelys Thalassemydidae Testudines Reptilia Chordata 128349 65907 56475 36322 33815
Aspideretoides Trionychidae Testudines Reptilia Chordata 67335 37674 56475 36322 33815
Judithemys Sinemydidae Testudines Reptilia Chordata 56471 37757 56475 36322 33815
<NA> Trionychidae Testudines Reptilia Chordata NA 37674 56475 36322 33815
Taphrosphys Bothremydidae Testudines Reptilia Chordata 37608 67318 56475 36322 33815
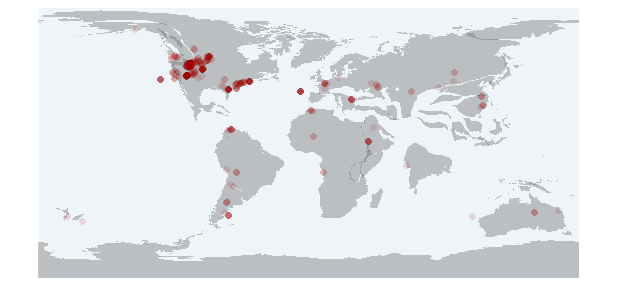
pm_plot
Returns a plot with the paleomap and the fossil occurrences.
data <- pm_getdata (base_name="Testudines",
interval="Paleocene")
pm_plot (interval="Paleocene", data)
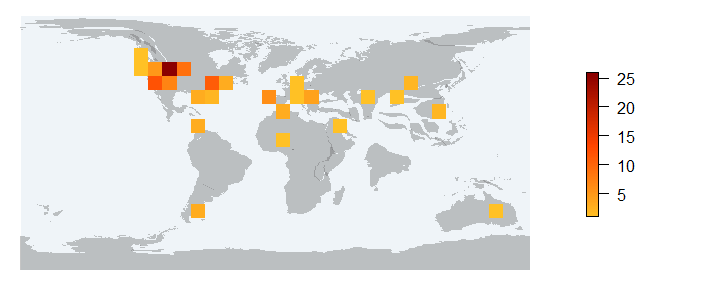
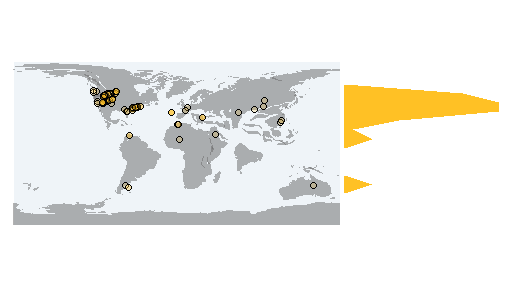
pm_occraster Returns a RasterLayer of the sampling effort and a map with the raster on it.
> shape <- pm_getmap(interval="Paleocene")
> data <- pm_getdata (base_name="Testudines",
interval="Paleocene")
> pm_occraster (shape, data, rank="species")
class : RasterLayer
dimensions : 18, 36, 648 (nrow, ncol, ncell)
resolution : 10, 10 (x, y)
extent : -180, 180, -92.0605, 87.9395 (xmin, xmax, ymin, ymax)
coord. ref. : NA
data source : in memory
names : layer
values : 1, 26 (min, max)
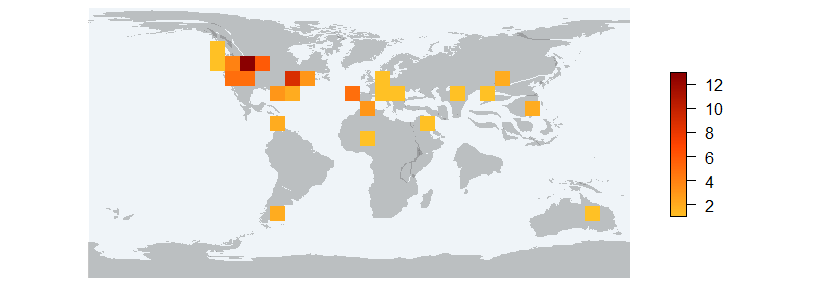
pm_richraster Returns a RasterLayer of richness and a map with the raster on it.
> pm_richraster (shape, data, rank="species")
class : RasterLayer
dimensions : 18, 36, 648 (nrow, ncol, ncell)
resolution : 10, 10 (x, y)
extent : -180, 180, -92.0605, 87.9395 (xmin, xmax, ymin, ymax)
coord. ref. : NA
data source : in memory
names : layer
values : 1, 13 (min, max)
pm_occ Returns a dataframe of taxa occurrences vs. localities (~ sites per taxa matrix)
> occ<- pm_occ (data, rank="species")
> str (occ)'data.frame': 75 obs. of 54 variables:
$ paleolat : num 40.1 38.8 40.9 53.1 10.1 ...
$ paleolng : num -44.5 -47.6 -39.8 -78.2 42.8 ...
$ Peritresius ornatus : num 1 0 1 0 0 0 0 0 0 0 ...
$ Aspideretoides virginianus: num 0 1 0 0 0 0 0 0 0 0 ...
$ Agomphus pectoralis : num 0 0 1 0 0 0 0 0 0 0 ...
$ Osteopygis emarginatus : num 0 0 1 0 0 0 0 0 0 0 ...
$ Cardichelyon rogerwoodi : num 0 0 0 1 0 0 0 0 0 0 ...
$ Arabemys crassiscutata : num 0 0 0 0 1 0 0 0 0 0 ...
$ Protochelydra zangerli : num 0 0 0 0 0 1 0 0 0 0 ...
$ Compsemys victa : num 0 0 0 0 0 0 1 2 1 1 ...
$ Taphrosphys sulcatus : num 0 0 0 0 0 0 0 0 0 0 ...
$ Agomphus oxysternum : num 0 0 0 0 0 0 0 0 0 0 ...
$ Bothremys maghrebiana : num 0 0 0 0 0 0 0 0 0 0 ...
$ Taphrosphys ippolitoi : num 0 0 0 0 0 0 0 0 0 0 ...
$ Rhothonemys brinkmani : num 0 0 0 0 0 0 0 0 0 0 ...
$ Agomphus alabamensis : num 0 0 0 0 0 0 0 0 0 0 ...
$ Cerrejonemys wayuunaiki : num 0 0 0 0 0 0 0 0 0 0 ...
$ Acleistochelys maliensis : num 0 0 0 0 0 0 0 0 0 0 ...
$ Yaminuechelys maior : num 0 0 0 0 0 0 0 0 0 0 ...
$ Naiadochelys patagonica : num 0 0 0 0 0 0 0 0 0 0 ...
$ Palatobaena bairdi : num 0 0 0 0 0 0 0 0 0 0 ...
$ Cedrobaena putorius : num 0 0 0 0 0 0 0 0 0 0 ...
$ Euclastes acutirostris : num 0 0 0 0 0 0 0 0 0 0 ...
$ Eosphargis insularis : num 0 0 0 0 0 0 0 0 0 0 ...
$ Osteopygis roundsi : num 0 0 0 0 0 0 0 0 0 0 ...
$ Tasbacka ruhoffi : num 0 0 0 0 0 0 0 0 0 0 ...
$ Mongolemys trufanensis : num 0 0 0 0 0 0 0 0 0 0 ...
$ Labrostochelys galkini : num 0 0 0 0 0 0 0 0 0 0 ...
$ Alamosemys substricta : num 0 0 0 0 0 0 0 0 0 0 ...
$ Hoplochelys crassa : num 0 0 0 0 0 0 0 0 0 0 ...
$ Conchochelys admirabilis : num 0 0 0 0 0 0 0 0 0 0 ...
$ Axestemys puercensis : num 0 0 0 0 0 0 0 0 0 0 ...
$ Puentemys mushaisaensis : num 0 0 0 0 0 0 0 0 0 0 ...
$ Berruchelus russelli : num 0 0 0 0 0 0 0 0 0 0 ...
$ Judithemys backmani : num 0 0 0 0 0 0 0 0 0 0 ...
$ Hutchemys rememdium : num 0 0 0 0 0 0 0 0 0 0 ...
$ Ronella botanica : num 0 0 0 0 0 0 0 0 0 0 ...
$ Paramongolemys khosatzkyi : num 0 0 0 0 0 0 0 0 0 0 ...
$ Elkemys australis : num 0 0 0 0 0 0 0 0 0 0 ...
$ Hokouchelys chenshuensis : num 0 0 0 0 0 0 0 0 0 0 ...
$ Axestemys montinsana : num 0 0 0 0 0 0 0 0 0 0 ...
$ Tasbacka ouledabdounensis : num 0 0 0 0 0 0 0 0 0 0 ...
$ Tasbacka aldabergeni : num 0 0 0 0 0 0 0 0 0 0 ...
$ Catapleura repanda : num 0 0 0 0 0 0 0 0 0 0 ...
$ Judithemys kranzi : num 0 0 0 0 0 0 0 0 0 0 ...
$ Trionyx halophilus : num 0 0 0 0 0 0 0 0 0 0 ...
$ Atoposemys entopteros : num 0 0 0 0 0 0 0 0 0 0 ...
$ Tullochelys montana : num 0 0 0 0 0 0 0 0 0 0 ...
$ Stygiochelys estesi : num 0 0 0 0 0 0 0 0 0 0 ...
$ Mongolemys reshetovi : num 0 0 0 0 0 0 0 0 0 0 ...
$ Chelodina alanrixi : num 0 0 0 0 0 0 0 0 0 0 ...
$ Derrisemys sterea : num 0 0 0 0 0 0 0 0 0 0 ...
$ Plastomenoides tetanetron : num 0 0 0 0 0 0 0 0 0 0 ...
$ Plastomenoides lamberti : num 0 0 0 0 0 0 0 0 0 0 ...pm_occ_cell Returns a dataframe of taxa occurrences vs. cells (aggregation of taxa based on cells and not on localities like pm_occ)
> occ_cell<- pm_occ_cell (data, rank="species", res=10)
> str (occ_cell)'data.frame': 648 obs. of 54 variables:
$ long : num -175 -165 -155 -145 -135 -125 -115 -105 -95 -85 ...
$ lat : num -85 -85 -85 -85 -85 -85 -85 -85 -85 -85 ...
$ Peritresius ornatus : num 0 0 0 0 0 0 0 0 0 0 ...
$ Aspideretoides virginianus: num 0 0 0 0 0 0 0 0 0 0 ...
$ Agomphus pectoralis : num 0 0 0 0 0 0 0 0 0 0 ...
$ Osteopygis emarginatus : num 0 0 0 0 0 0 0 0 0 0 ...
$ Cardichelyon rogerwoodi : num 0 0 0 0 0 0 0 0 0 0 ...
$ Arabemys crassiscutata : num 0 0 0 0 0 0 0 0 0 0 ...
$ Protochelydra zangerli : num 0 0 0 0 0 0 0 0 0 0 ...
$ Compsemys victa : num 0 0 0 0 0 0 0 0 0 0 ...
$ Taphrosphys sulcatus : num 0 0 0 0 0 0 0 0 0 0 ...
$ Agomphus oxysternum : num 0 0 0 0 0 0 0 0 0 0 ...
$ Bothremys maghrebiana : num 0 0 0 0 0 0 0 0 0 0 ...
$ Taphrosphys ippolitoi : num 0 0 0 0 0 0 0 0 0 0 ...
$ Rhothonemys brinkmani : num 0 0 0 0 0 0 0 0 0 0 ...
$ Agomphus alabamensis : num 0 0 0 0 0 0 0 0 0 0 ...
$ Cerrejonemys wayuunaiki : num 0 0 0 0 0 0 0 0 0 0 ...
$ Acleistochelys maliensis : num 0 0 0 0 0 0 0 0 0 0 ...
$ Yaminuechelys maior : num 0 0 0 0 0 0 0 0 0 0 ...
$ Naiadochelys patagonica : num 0 0 0 0 0 0 0 0 0 0 ...
$ Palatobaena bairdi : num 0 0 0 0 0 0 0 0 0 0 ...
$ Cedrobaena putorius : num 0 0 0 0 0 0 0 0 0 0 ...
$ Euclastes acutirostris : num 0 0 0 0 0 0 0 0 0 0 ...
$ Eosphargis insularis : num 0 0 0 0 0 0 0 0 0 0 ...
$ Osteopygis roundsi : num 0 0 0 0 0 0 0 0 0 0 ...
$ Tasbacka ruhoffi : num 0 0 0 0 0 0 0 0 0 0 ...
$ Mongolemys trufanensis : num 0 0 0 0 0 0 0 0 0 0 ...
$ Labrostochelys galkini : num 0 0 0 0 0 0 0 0 0 0 ...
$ Alamosemys substricta : num 0 0 0 0 0 0 0 0 0 0 ...
$ Hoplochelys crassa : num 0 0 0 0 0 0 0 0 0 0 ...
$ Conchochelys admirabilis : num 0 0 0 0 0 0 0 0 0 0 ...
$ Axestemys puercensis : num 0 0 0 0 0 0 0 0 0 0 ...
$ Puentemys mushaisaensis : num 0 0 0 0 0 0 0 0 0 0 ...
$ Berruchelus russelli : num 0 0 0 0 0 0 0 0 0 0 ...
$ Judithemys backmani : num 0 0 0 0 0 0 0 0 0 0 ...
$ Hutchemys rememdium : num 0 0 0 0 0 0 0 0 0 0 ...
$ Ronella botanica : num 0 0 0 0 0 0 0 0 0 0 ...
$ Paramongolemys khosatzkyi : num 0 0 0 0 0 0 0 0 0 0 ...
$ Elkemys australis : num 0 0 0 0 0 0 0 0 0 0 ...
$ Hokouchelys chenshuensis : num 0 0 0 0 0 0 0 0 0 0 ...
$ Axestemys montinsana : num 0 0 0 0 0 0 0 0 0 0 ...
$ Tasbacka ouledabdounensis : num 0 0 0 0 0 0 0 0 0 0 ...
$ Tasbacka aldabergeni : num 0 0 0 0 0 0 0 0 0 0 ...
$ Catapleura repanda : num 0 0 0 0 0 0 0 0 0 0 ...
$ Judithemys kranzi : num 0 0 0 0 0 0 0 0 0 0 ...
$ Trionyx halophilus : num 0 0 0 0 0 0 0 0 0 0 ...
$ Atoposemys entopteros : num 0 0 0 0 0 0 0 0 0 0 ...
$ Tullochelys montana : num 0 0 0 0 0 0 0 0 0 0 ...
$ Stygiochelys estesi : num 0 0 0 0 0 0 0 0 0 0 ...
$ Mongolemys reshetovi : num 0 0 0 0 0 0 0 0 0 0 ...
$ Chelodina alanrixi : num 0 0 0 0 0 0 0 0 0 0 ...
$ Derrisemys sterea : num 0 0 0 0 0 0 0 0 0 0 ...
$ Plastomenoides tetanetron : num 0 0 0 0 0 0 0 0 0 0 ...
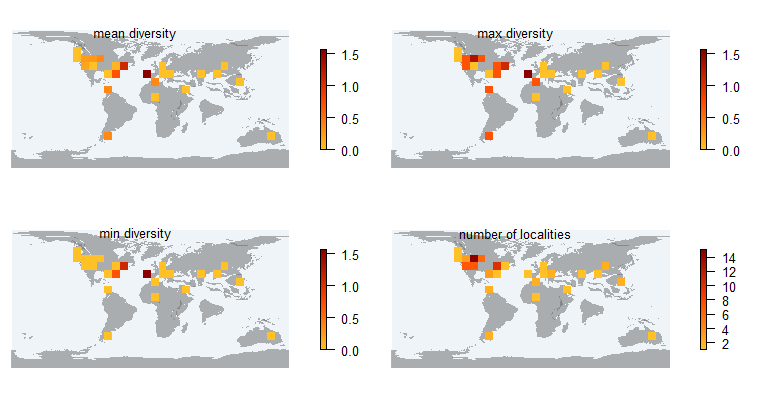
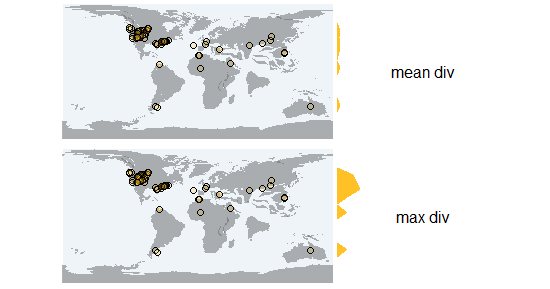
$ Plastomenoides lamberti : num 0 0 0 0 0 0 0 0 0 0 ...pm_divraster_loc 1- calculates the Shannon diversity per unique locality (based on its coordinates), 2- makes a raster file and a plot showing mean, max, min diversity per cell, or number of unique localities per cell
> occ_df <- pm_occ (data, rank="species")
> pm_divraster_loc (shape, occ_df, fun=mean)
> pm_divraster_loc (shape, occ_df, fun=max)
> pm_divraster_loc (shape, occ_df, fun=min)
> pm_divraster_loc (shape, occ_df, fun="count")
class : RasterLayer
dimensions : 18, 36, 648 (nrow, ncol, ncell)
resolution : 10, 10 (x, y)
extent : -180, 180, -92.0605, 87.9395 (xmin, xmax, ymin, ymax)
coord. ref. : NA
data source : in memory
names : layer
values : 1, 15 (min, max)
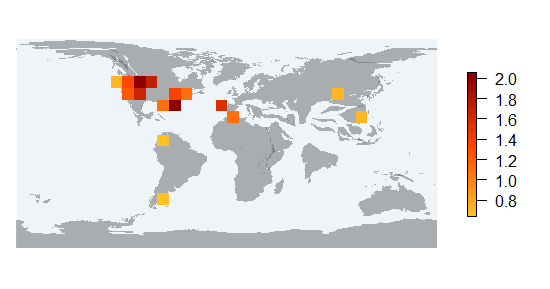
pm_divraster_cell calculates the Shannon diversity per cell (taking into account relative abundances of all the fossil records whithin the cell)
occ_df_cell <- pm_occ_cell (data, rank="species")
pm_divraster_cell (shape, occ_df_cell, res=10)class : RasterLayer
dimensions : 18, 36, 648 (nrow, ncol, ncell)
resolution : 10, 10 (x, y)
extent : -180, 180, -92.0605, 87.9395 (xmin, xmax, ymin, ymax)
coord. ref. : NA
data source : in memory
names : layer
values : 0.6365142, 2.059229 (min, max)pm_latrich calculates latitudinal diversity of taxa (species, genera, families, orders)
> pm_latrich (shape, data, rank="species", res=10) lat_min lat_max richn
1 -90 -80 0
2 -80 -70 0
3 -70 -60 0
4 -60 -50 0
5 -50 -40 3
6 -40 -30 0
7 -30 -20 0
8 -20 -10 0
9 -10 0 0
10 0 10 3
11 10 20 1
12 20 30 6
13 30 40 17
14 40 50 17
15 50 60 13
16 60 70 0
17 70 80 0
18 80 90 0pm_latdiv
calculates the Shannon diversity along the latitudinal gradient based on the individual values of diverstiy the fossil localities of those latitudes. The function returns the mean or max values of diversity of the sampled localities along the latitudinal gradient.
> occ_df <- pm_occ (data)
> pm_latdiv (occ_df, shape, fun=mean)
> pm_latdiv (occ_df, shape, fun=max)
maxlat minlat div
1 -90 -80 0.0000000
2 -80 -70 0.0000000
3 -70 -60 0.0000000
4 -60 -50 0.0000000
5 -50 -40 0.6931472
6 -40 -30 0.0000000
7 -30 -20 0.0000000
8 -20 -10 0.0000000
9 -10 0 0.0000000
10 0 10 0.6931472
11 10 20 0.0000000
12 20 30 0.6931472
13 30 40 1.5607104
14 40 50 1.3321790
15 50 60 1.0986123
16 60 70 0.0000000
17 70 80 0.0000000
18 80 90 0.0000000Please report any issues or bugs.
License: GPL-2
To cite package paleoMap in publications use:
To cite package `paleoMap` in publications use:
Sara Varela, K. Sonja Rothkugel (2015). paleoMap: combine paleogeography and paleobiodiversity. R package version 0.1. https://github.com/macroecology/paleoMap
A BibTeX entry for LaTeX users is
@Manual{,
title = {paleoMap: combine paleogeography and paleobiodiversity},
author = {Sara Varela} and {Sonja Rothkugel},
year = {2016},
note = {R package version 0.1},
base = {https://github.com/macroecology/paleoMap},
}