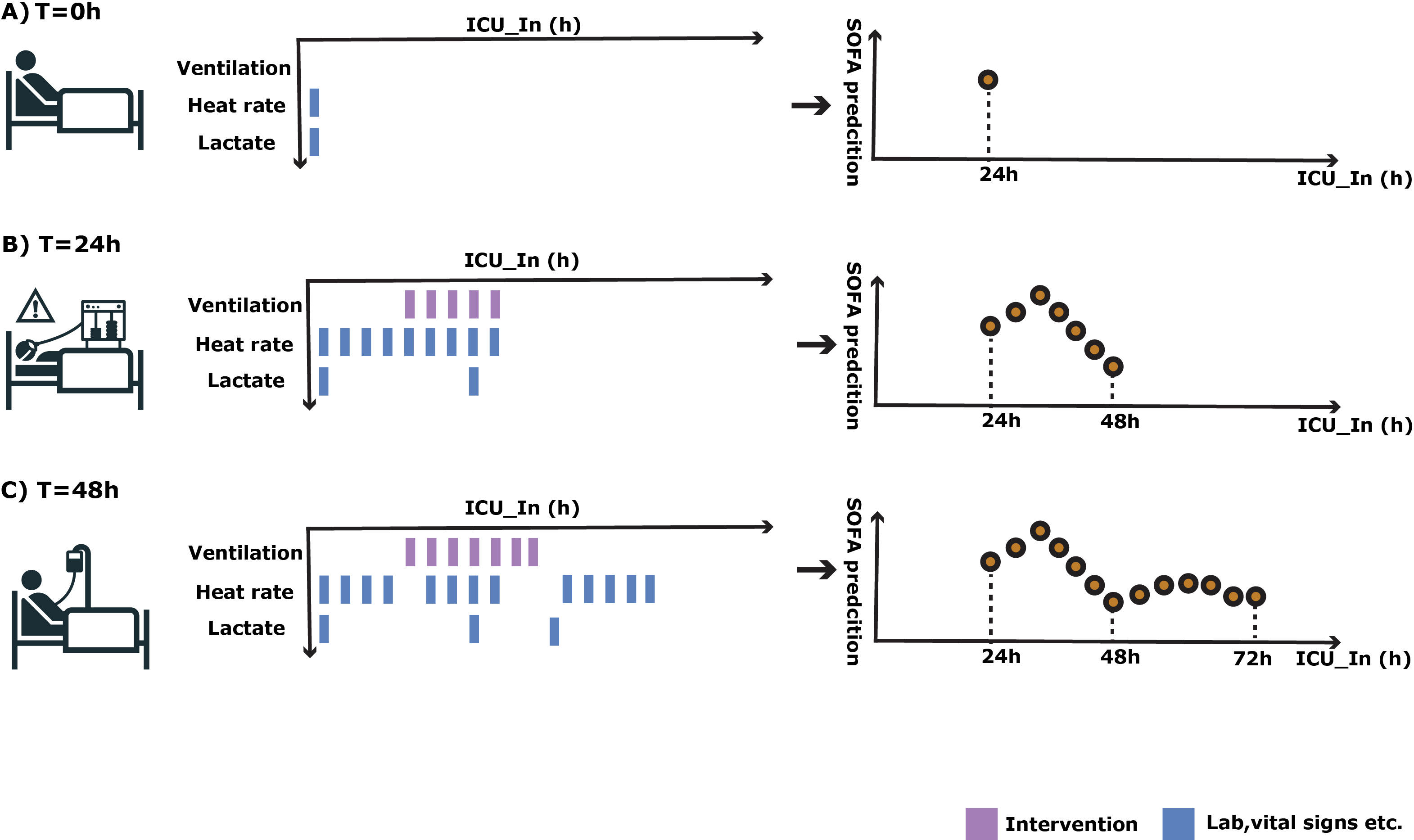
Predict 24h later SOFA score using MIMIC-IV and eICU data
python main.py --model_name TCN --num_channels 256 256 256 256 --kernel_size 3 --checkpoint test
python main.py --model_name Transformer --d_model 256 --n_head 8 --dim_ff_mul 4 --num_enc_layer 2 --checkpoint test
python main.py --model_name RNN --hidden_dim 256 --layer_dim 3 --checkpoint test
To use sepsis-3 cohort: do
python main.py --use_sepsis3 --model_name TCN --num_channels 256 256 256 256 --kernel_size 3 --checkpoint test
Different ways of data batching: same, close, random. Default is close,
Same: each minibatch has the same length of records. This sets an upper limit on the batch size since length 216 records are not common in MIMIC-IV.
Close: each minibatch has very close length of records. This way data padding is minimal. The parameter to set how close they are is bucket_size default is 300. The smaller this parameter, the closer of the length in each minibatch.
Random: random minibatch. Under this, each batch could have mixture of length 1 and length 200 records.
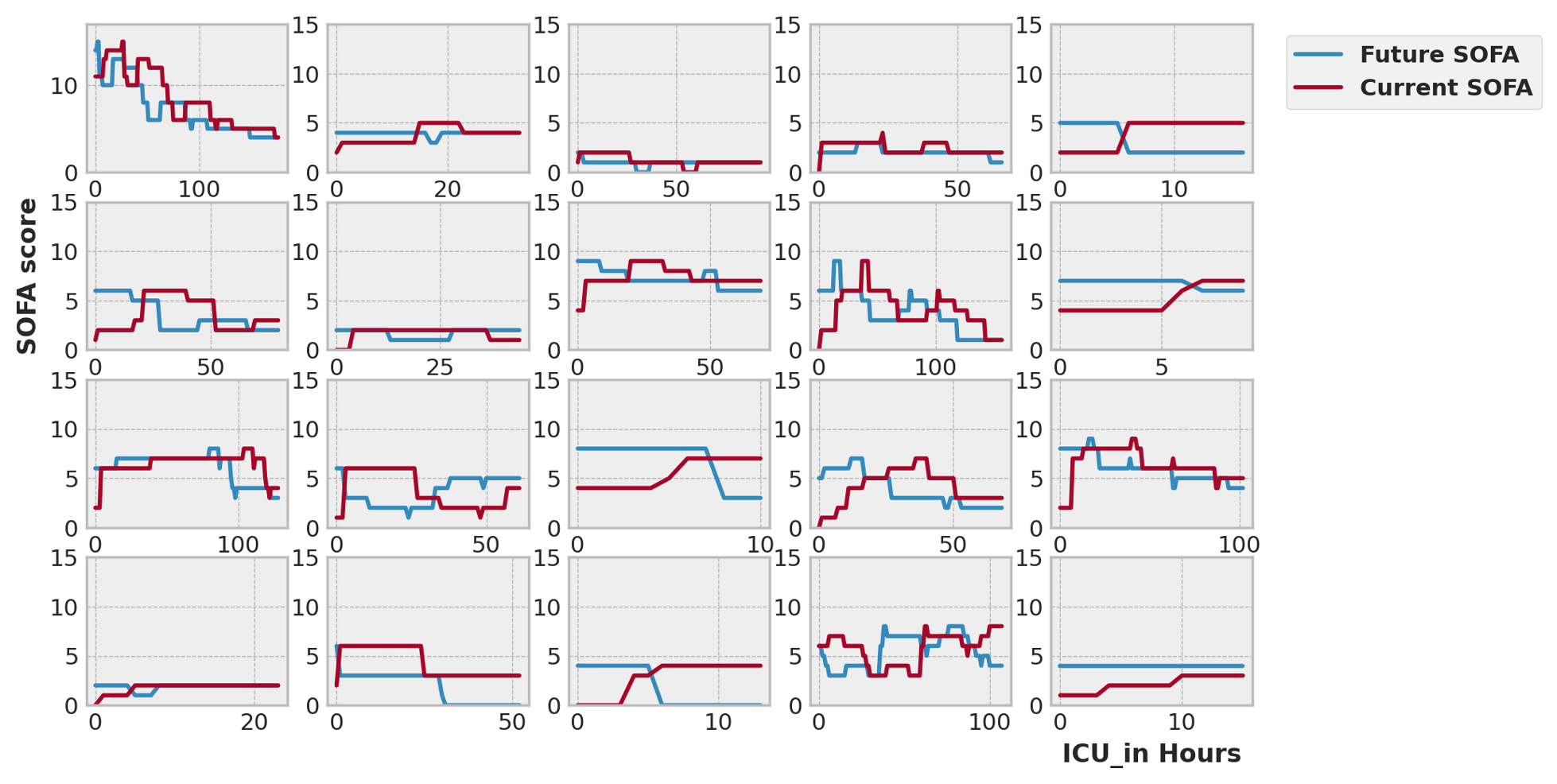
If we use the naive hypothesis, under which patient SOFA scores don't change during 24h, we get a huge error (MSE: 6.34). As can be seen in the following randomly selected 20 records.
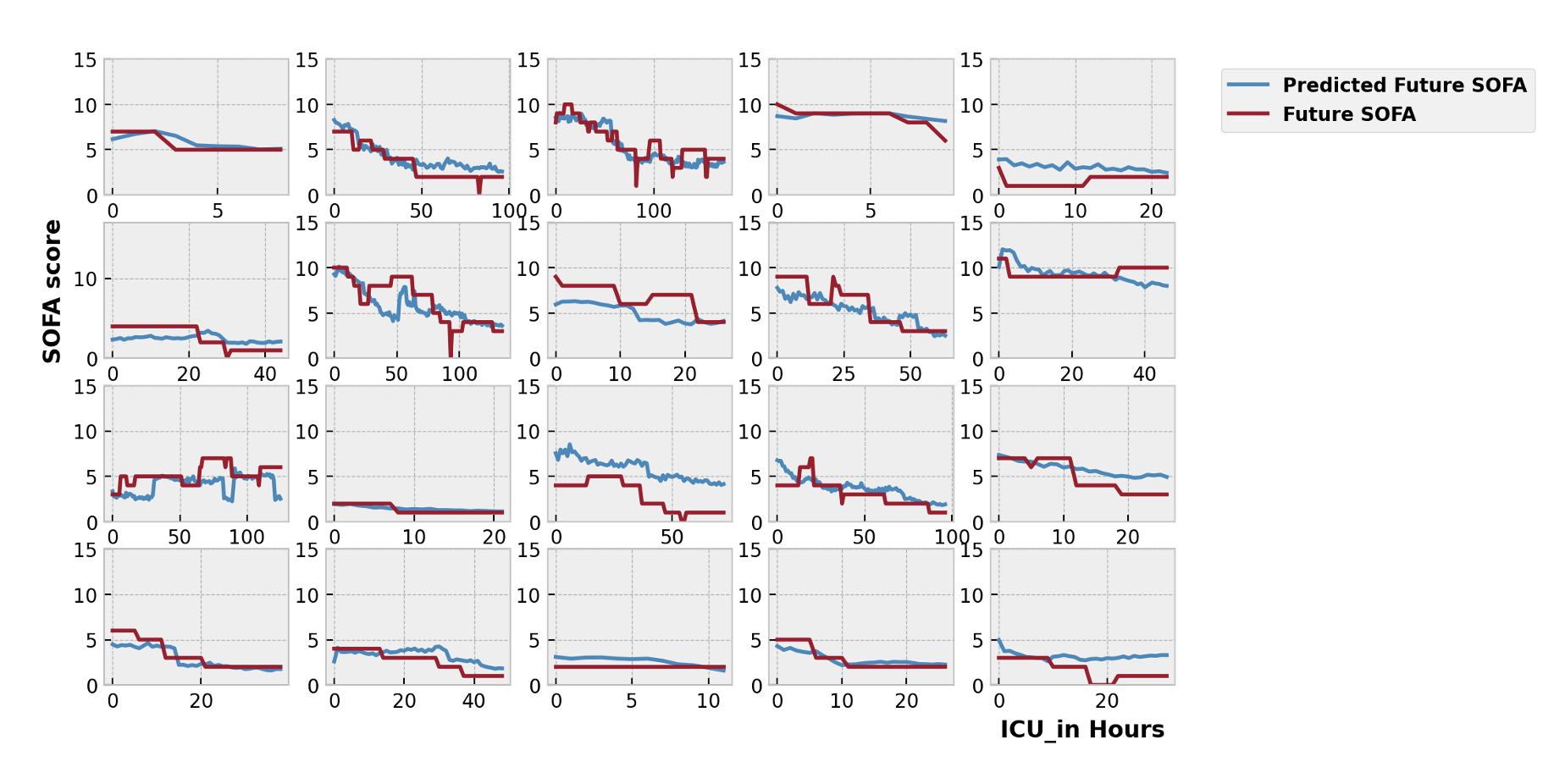
If we use the TCN model, we can see it predicts the rapidly changing trends (MSE: 2.54).