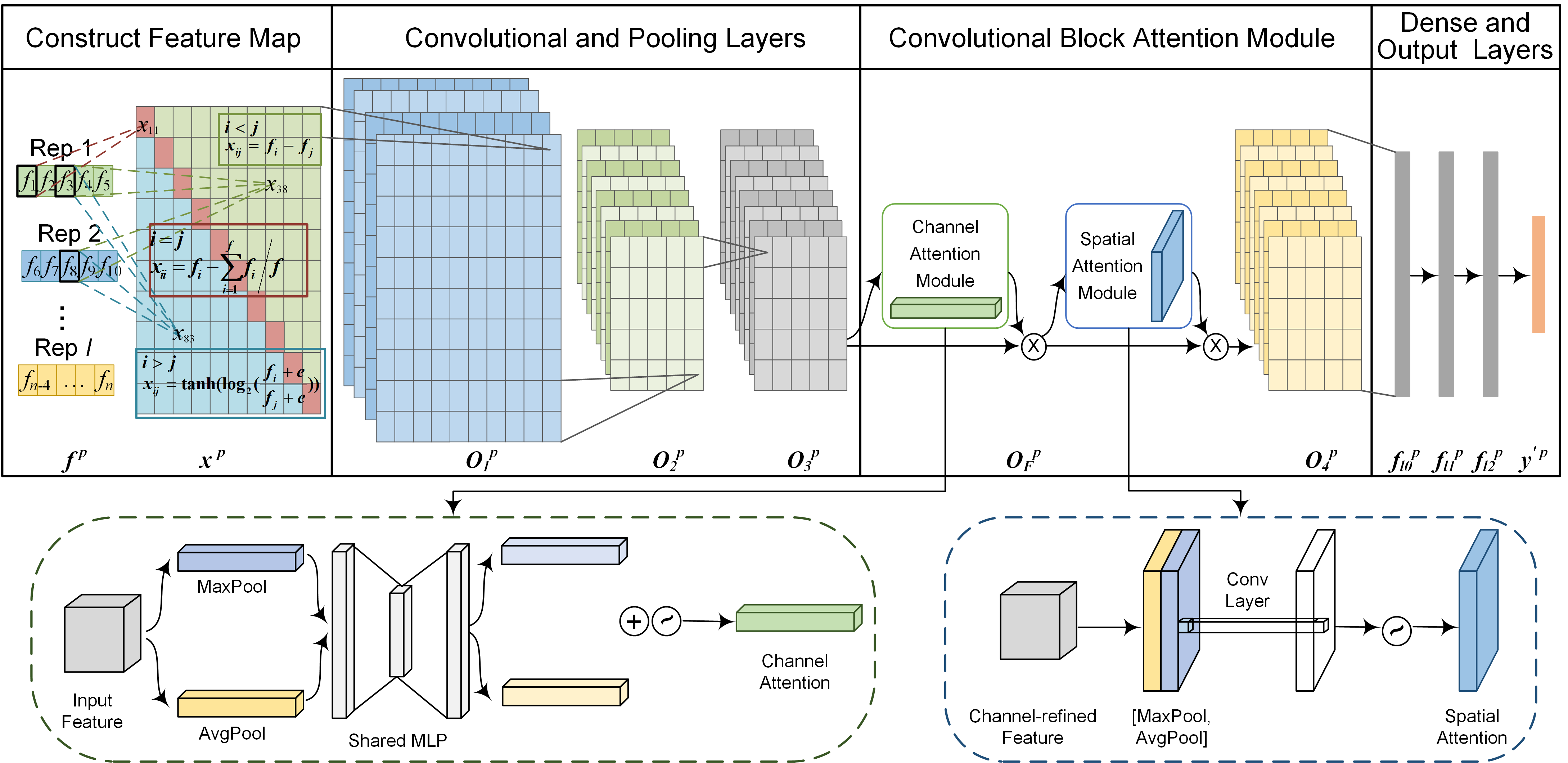
DeepSP: A Deep Learning Framework for Spatial Proteomics
DeepSP.py is is a python script that requires python software and some python modules to be installed.
The python modules that need to be installed are listed in requirements.txt, they can be installed via the command line interface as follows
$ pip install -r requirements.txtIf any python modules fail to install, you can install them one by one with the following command
$ pip install pandas
$ pip install numpy
$ pip install sklearn
$ pip install argparse
$ pip install time
$ pip install copy
$ pip install os
$ pip install warnings
$ pip install conda install pytorch torchvision torchaudio cpuonly -c pytorch
# or
$ pip3 install torch torchvision torchaudioThe introduction of the DeepSP parameters can be achieved by naming them as follows
$ python DeepSP.py -h
usage: DeepSP.py [-h] [-f F] [-r R] [-p {Y,N}] [-n N] [-bs BS] [-s S] [-lr LR] [-l2 L2] [-eps EPS] [-nep NEP]
DeepSP for Protein Subcellular Localization Prediction
optional arguments:
-h, --help show this help message and exit
-f F, --file F file path, first column must be ProteinID, last column must be markers
-r R, --rep R number of repeated experiments
-p {Y,N}, --getpred {Y,N}
whether to predict unknown proteins, default=Y
-n N, --n_splits N n-fold cross validation, default=5
-bs BS, --batch_size BS
mini batch size, default=64
-s S, -seed S a random seed, default=0
-lr LR, --learning_rate LR
learning rate, default=1e-3
-l2 L2, --l2_regularization L2
l2 regularization, default=1e-4
-eps EPS, --epochs EPS
training times, default=100
-nep NEP, --nepoch NEP
print the results every * times, default=20hirst2018.csv The file must be in csv format, the first column must be the protein number and there must be no duplicates, the last column must be the subcellular organelle to which the protein is localised, unknown localised proteins should be marked as unkonwn, the data in each row cannot be all zeros.
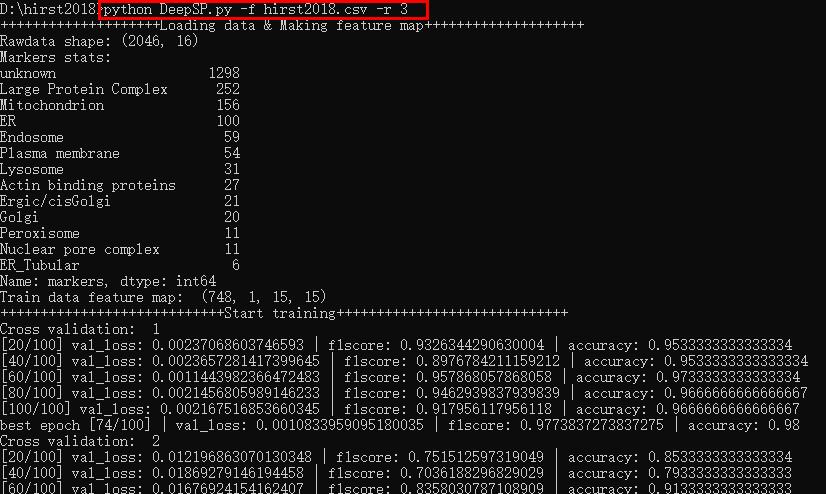
DeepSP.py can be run from the command line interface with the following commands, where -f (dataset file) and -r (the number of replicates) are the two required parameters. An example command is “python DeepSP.py -f hirst2018.csv -r 3”, in which “-f” is input dataset file and “-r” is the number of replicates. There is a running example in the example.jpg image。
$ python DeepSP.py -f hirst2018.csv -r 3