The cell phenotyping framework presented here is just for review purposes.
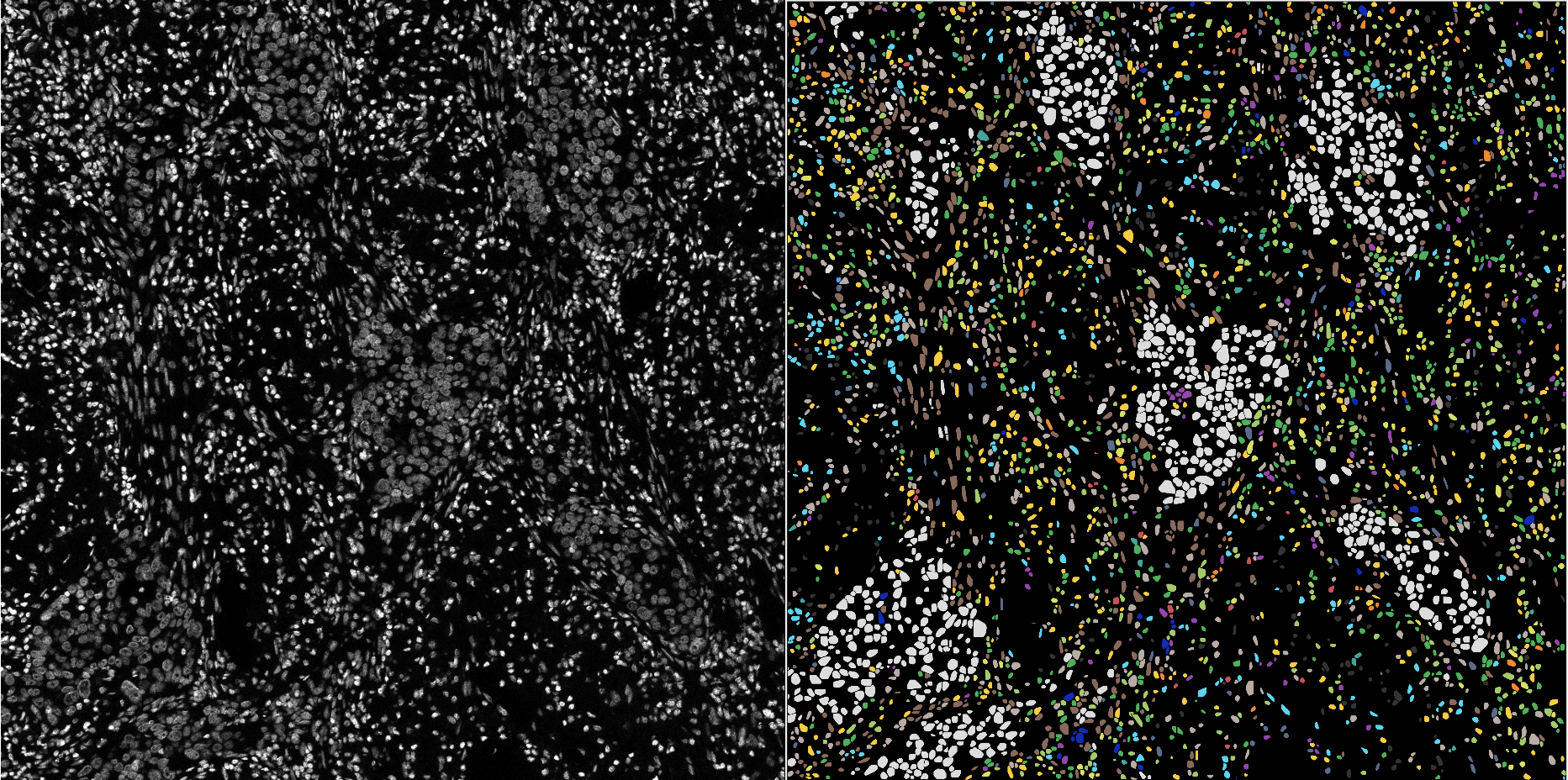
An example of cell phenotyping on a complex cohort. Please note that unidentified cells are plotted in dark gray.
To be able to run the cell segmentation framework, one needs the following:
To be able to run the compiler function for cell phenotyping compile_rules.m, the user should specify the following parameters in the config file (within the config folder. The currently used config file in the compile_rules.m is set to use the config_template.xlsx. In the config_template.xlsx, there are the following parameters to be set:
input_pano_folder: the folder where the raw images are located at.input_cell_segmentation_folder: the folder where the cell segmentation results are located at. This folder will have the same file organization (structure) as the input, given that the cell segmentation is done using our pipeline. For the cell segmentation pipeline, please refer to the GitHub repository located here: https://github.com/cellfactory411/Cell-Segmentationinput_keyPatientID: this is the address for a spreadsheet that has keys and patient IDs. The spreadsheet should contain two columnsFileNameandUniqueImageID. Generally, we have a one-to-one mapping between filenames and unique image ids but there could exist cases where we have multiple cores of one unique image id and this is predicted by our software and handled at the deeper level of the software.group_PatientID: this is the address for a spreadsheet that the groups and patient ids relationships. one other input to our code is that we can image multiple groups for our analysis. These groups can be coming from biological experimental designs and be easily input into the code. If there are no specific groups for your analysis, one can create just a single group with all the patient IDs in the spreadsheet.output_folder: this is where the output cell types are stored. Please note that this code produces two types of output. One is a folder where the PDF figures of cell-type assignments are produced and the second one is a.matfile that includes a data structure calledcellTypeswhere each index of that is a one-to-one map to the segmentation result, i. e., the n-th cell segmented in the segmentation folder gets its type in the n-th row in the mat data structure.IndicatorsListPath: this is the address to where the indictors list is saved as a spreadsheet.nuc_csv_source_folder&ring_csv_source_folder: this is the address to the folder where the code precomputes the intensities within each nucleus and its surroundings. The main idea here is that, if for some reason, the cell type assignment process is halted, the code recovers the intensities precomputed from these folders. Thus, these folders can be initialized to empty folders at the beginning.rule_table_location_xxxx: the address to the rule tables. We have uploaded an example rule table within this repository which is used for cell type assignment. This rule table is read and compiled by the code automatically. We have some tuning nobes for sensitivity which can be used to get the best possible results.